Automated HER2 testing: Personalized healthcare for breast cancer patients enabled by novel molecular morphology methods
Breast cancer is the second leading cause of cancer death in women in the United States. Copy number of the HER2 (human epidermal growth factor receptor 2) oncogene located on chromosome 17 is increased in approximately 18% to 20% of breast cancer patients. Amplification of the HER2 gene results in over-expression of HER2 protein on the cell membrane. In normal cells, there are two copies of the HER2 gene in each cell, with the HER2 cell signaling pathway normally controlling cell growth and division. However, as the HER2 gene is multiplied in tumor cells, over-expression of HER2 protein in tumor cells drives unregulated cell growth as the primary basis for carcinogenesis. HER2 positivity is associated with poor prognosis for breast cancer patients.
Personalized healthcare allows the tailoring of patient treatments based on patients’ genomic information. The U.S. Food and Drug Administration (FDA) has approved two antibody-based drugs for HER2 targeted therapy for treating HER2 positive breast cancer patients: 1) a humanized monoclonal anti-HER2 antibody, trastuzumab (Herceptin), in 1998; and 2) trastuzumab conjugated with a cytotoxic agent, emtansine (Kadcyla), in 2013. Binding of a therapeutic HER2 antibody to the extracellular domain of HER2 protein in tumor cells causes a reduction in cell proliferation and tumor shrinkage through a series of mechanisms. HER2 amplification and over-expression in breast cancer are predictive of response to HER2 targeted therapies, and HER2 testing has become standard of care as companion diagnostic assays. However, optimal usage of these drugs from a health economic and drug safety perspective is dependent on highly accurate and reproducible methods for HER2 assessment in the routine pathology laboratory environment. Manual and laboratory-developed methods demonstrate variable accuracy and reproducibility across laboratories in comparison with automated in vitro diagnostic (IVD) assays. Given the potential significant negative health consequences of false-positive and false-negative companion diagnostic assays, patient safety is best served by the development of standardized, automated IVD companion diagnostic assays for the many targeted therapies currently in drug development. This article will describe the HER2 assays currently available for routine laboratory testing with a focus on novel automated methods.
The American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) published the guidelines for HER2 testing in 2007 with a goal of improving the accuracy of HER2 testing in invasive breast cancer and its utility as a predictive marker.1 These guidelines included specific recommendations for HER2 immunohistochemistry (IHC) as well as in situ hybridization (ISH) in formalin-fixed, paraffin-embedded (FFPE) tissues. The FDA approved three HER2 IHC diagnostic assays using mouse monoclonal, rabbit polyclonal, or rabbit monoclonal anti-HER2 antibodies in 2012, 1998, and 2007, respectively. An independent quality assurance organization, NordiQC, reported that the HER2 IHC assay with a rabbit monoclonal anti-HER2 antibody (clone 4B5) was the most reliable method for HER2 protein status assessment.2
HER2 IHC assays are scored using a 3+, 2+, 1+, or negative scale based on IHC membrane intensity. Breast cancer patients with HER2 IHC staining of 3+ are considered HER2 positive, and eligible for a HER2 targeted therapy. Breast cancer patients with HER2 IHC staining of 2+ are categorized as HER2 equivocal, and their tissue samples should be tested with a reflex HER2 ISH assay. Breast cancer patients with 1+ or negative results are not candidates for HER2 targeted therapy. The FDA approved two dark-field HER2 ISH assays (fluorescence ISH) and three bright-field HER2 ISH assays (chromogenic ISH) for assessing HER2 gene status in breast cancer patients.
There are two types of bright-field chromogenic HER2 ISH assays: 1) single color ISH for the HER2 gene only; and 2) dual color ISH for the HER2 gene and CEN17 (chromosome 17 centromere). With the single color bright-field HER2 ISH assay, an average of six or more HER2 signals per nucleus are considered HER2 positive per ASCO/CAP guidelines, with four-six HER2 signals considered equivocal and less than four signals considered HER2 negative. With a dual color bright-field HER2 ISH assay, HER2 status is determined with the ratio of HER2 gene copy numbers to CEN17 copy numbers (negative: HER2/CEN17 ratio <1.8; equivocal: HER2/CEN17 ratio 1.8-2.2; positive: HER2/CEN17 ratio >2.2). Further analyses of HER2 gene and CEN17 copy numbers are required for the equivocal cases.
In order to avoid misinterpreting chromosome 17 polysomy as HER2 gene amplification, the use of a CEN17 probe is recommended for more accurate HER2 status assessment.3 Thus, a dual color ISH assay for HER2 gene and CEN17 targets is especially suitable for HER2 equivocal cases and chromosome 17 polysomy cases. It should be noted that pre-analytical conditions such as fixation state can affect the performance of both HER2 IHC and ISH assays. With HER2 ISH assays HER2 signals in normal cells adjacent to the tumor can be used as convenient built-in normal controls to ensure proper assay sensitivity.
HER2 fluorescence ISH assays can be achieved by manual, semi-automated, or fully automated protocols. There are several disadvantages associated with fluorescence ISH applications: 1) the necessity of a specialized and expensive fluorescence microscope; 2) the assay must be interpreted in a dark room; 3) the problem of fluorescent signal quenching and inability to preserve ISH signals for long-term storage; and 4) the poor cellular and tissue morphology associated with FISH can make it challenging to localize signals to appropriate regions of the sample (e.g., invasive tumor areas). Furthermore, if a traditional manual protocol for HER2 fluorescence ISH assay is used, it can introduce human errors during a multi-step two-day process. The use of an automated ISH protocol is recommended for reproducible results.
HER2 chromogenic ISH applications are also manual, semi-automated, or fully automated protocols. Chromogenic ISH assays have several advantages: 1) the assay is interpreted via a standard light microscope; 2) the photo-stability of chromogenic ISH signals enable re-interpretation after long-term storage; and 3) hematoxylin counterstaining creates excellent tissue morphology and improved ability to correlate ISH signals with specific tissue areas. The NordiQC quality assurance organization conducted comparison studies between fluorescence-based HER2 ISH and chromogenic-based HER2 ISH applications and concluded that commercially available chromogenic HER2 ISH assays can be used for diagnosing the HER2 status of breast cancer.4
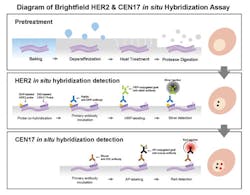
A technical paper describing the first generation of a fully automated, unique bright-field HER2 and CEN17 dual color ISH application was published in 2008.5 This assay was created by combining two kinds of hybridization and signal detection chemistries for HER2 gene and CEN17 targets into one assay on a single tissue section. In this method, hybridization and silver signal detection for the HER2 gene was followed by a hybridization and red signal detection step for CEN17.
A multicenter ring study was conducted to assess the assay performance of the HER2 Dual ISH application by The United Kingdom National External Quality Assessment Service for Immunocytochemistry (UK NEQAS), and a high concordance rate between the bright-field and dark-field HER2 assays for diagnosing HER2 status was demonstrated.6 A fully automated, bright-field HER2 and CEN17 dual color ISH application was approved by the FDA as well as the Japanese Ministry of Health & Welfare (JMHW) for the assessment of breast cancer patients considered for Herceptin therapy in 2011.
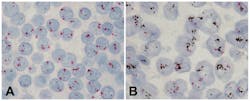
The current HER2 Dual ISH application displays HER2 gene and CEN17 as black dots and red dots, respectively. The technical process for the assay begins with baked, formalin-fixed, paraffin-embedded breast tissue sections being subjected to the removal of paraffin from tissue sections followed by pretreatment steps with heat treatment and protease digestion (Figure 1). HER2 gene and CEN17 targets are then localized with a cocktail of 2,4-dinitrophenyl (DNP)-labeled probe and digoxigenin (DIG)-labeled CEN17 probe. Finally, the HER2 gene and CEN17 are visualized with silver and red chromogens, respectively. The silver and red chromogens combined with a hematoxylin counterstain allow correlation of gene signals with cellular morphology (Figure 2). Multiple studies have compared HER2 Dual ISH assay performance to that of HER2 fluorescence ISH and HER2 IHC assays not only for breast cancer, but also for gastric cancer. The medical value of the assay has been presented as an alternative method for assessing the HER2 status of cancer patients.7-11
Automated bright-field ISH assays such as HER2 Dual ISH can free pathologists and medical technologists from complex, technically challenging manual protocols and represent a new generation of molecular assays that provide genomic and proteomic information in the context of traditional histomorphology. These “molecular morphology” assays will be critically important for continued progress in the areas of personalized healthcare, precision medicine, and companion diagnostics. As we continue to learn more about the complexity of cancer biology including acquired resistance mechanisms and intra/inter-tumor heterogeneity, it will be increasingly important for pathologists to utilize these types of assays to comprehensively profile tumors, enabling the matching of tumor driver mechanisms to the optimal selection of targeted therapies to maximize patient outcomes.
References
- Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118-145.
- Assessment Run B14 2012 HER-2 IHC. http://www.nordiqc.org/Run-36-B14-H2/Assessment/assessment-B14-HER2.htm. Accessed April 29, 2013.
- Gruver AM, Peerwani Z, Tubbs RR. Out of the darkness and into the light: bright field in situ hybridization for delineation of ERBB2 (HER2) status in breast cancer. J Clin Pathol. 2010;63:210-219.
- Assessment Run H2 2012 HER-2 ISH. http://www.nordiqc.org/Run-36-B14-H2/Assessment/assessment-H2-HER-2.htm. Accessed April 29, 2013.
- Nitta H, Hauss-Wegrzyniak B, Lehrkamp M, et al. Development of automated brightfield double in situ hybridization (BDISH) application for HER2 gene and chromosome 17 centromere (CEN17) for breast carcinomas and an assay performance comparison to manual dual color HER2 fluorescence in situ hybridization (FISH). Diagn Pathol. 2008;3:41.
- Bartlett JMS, Campbell FM, Ibrahim M, et al. A UK NEQAS ISH multicenter ring study using the Ventana HER2 dual-color ISH assay. Am J Clin Pathol. 2011;135:157-162.
- Koh YW, Lee HJ, Lee JW, Kang J, Gong Gyungyub. Dual-color silver-enhanced in situ hybridization for assessing HER2 gene amplification in breast cancer. Mod Pathol. 2011;24:794-800.
- Yan B, Yau EX, Choo SN, et al. Dual-colour HER2/chromosome 17 chromogenic in situ hybridisation assay enables accurate assessment of HER2 genomic status in gastric cancer and has potential utility in HER2 testing of biopsy samples. J Clin Pathol. 2011;64:880-883.
- Grin A, Brezden-Masley C, Bauer S, Streutker CJ. HER2 in situ hybridization in gastric and gastroesophageal adenocarcinoma: comparison of automated dual ISH to FISH. Appl Immunohistochem Mol Morphol. 2013 [Epub ahead of print]
- Horii R, Matsuura M, Iwase T, Ito Y, Akiyama F. Comparison of dual-color in-situ hybridization and fluorescence in-situ hybridization in HER2 gene amplification in breast cancer. Breast Cancer. 2013 [Epub ahead of print]
- Kosa G, Kardos L, Kovacs J, Szollosi Z. Comparison of dual-color dual-hapten brightfield in situ hybridization (DDISH) and fluorescence in situ hybridization in breast cancer HER2 assessment. Pathol Res Pract. 2013;209:147-150.