Genetic testing has been a part of the clinical diagnostic strategy for more than half a century. The first diagnostic test, a molecular-based phenylketonuria (PKU) newborn screening assay, detected levels of phenylalanine in the blood using a bacterial inhibition assay. Two decades later the first disease-associated gene, Huntingtin (HTT), was mapped, and restriction fragment length polymorphism (RFLP) analysis was the method of choice for tracing the transmission of mutant alleles.1 While some older techniques for genetic testing are still applicable today, the clinical implementation of DNA sequencing has revolutionized diagnostics.
During the last several years new sequencing technologies have significantly enhanced the identification of disease-associated mutations and discovery of causative genetic alterations. As a result, there are currently more than 2,500 genetic conditions with a commercially available diagnostic test.2 Despite these advancements, patient access to genetic testing for rare or orphan genetic disorders can be limited, depending on prevalence and number of genes involved. Many Mendelian disorders such as cystic fibrosis are caused by mutations in a single gene, whereas other inherited disorders, such as X-Linked Intellectual Disability (XLID), can have a significant number of underlying causative genes. Typically, testing for rare diseases is not cost-effective, as the samples cannot be batched for processing due to low volume. Therefore, most diagnostic labs focus on the more common disorders to limit costs. Traditionally, Sanger sequencing has been considered the gold standard in mutation detection and is still the method of choice for most diagnostic labs. However, this technology is not ideal for screening large sets of genes involved in molecularly heterogeneous disorders.
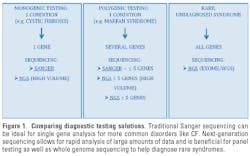
Recent advancements in sequencing technologies have made it possible to generate a large amount of data quickly and cost-effectively. By removing most of the throughput and resource constraints seen with traditional methods, next generation sequencing (NGS) provides investigators with required capacity to analyze large panels of genes or whole genomes in a single run. Not surprisingly, these transformative technologies are enabling new tailor-made approaches to diagnostic testing and providing solutions for even the rarest disorders (Figure 1).
Recently, Dr. Stephen Kingsmore’s team at Children’s Mercy Hospital in Kansas City, Missouri, utilized new sequencers to perform whole genome sequencing of newborns in neonatal intensive care. Due to the speed of the machines, a specialized bioinformatics pipeline, and no need for target enrichment they were able to identify the relevant mutations responsible for several of the infants’ symptoms within 50 hours.3 Although this is impressive, there are several limitations that will restrict whole genome sequencing to only the most severe and time-sensitive cases in the near term.
First, the cost of whole genome sequencing diagnostic service is significant, and involvement of major insurance providers in covering these costs has not yet been established. (Dr. Kingsmore’s group estimates the complete cost, including analysis, will be $13,500.)
Next, to perform whole genome sequencing in high volume, a diagnostic lab needs a considerable number of machines and amount of bioinformatics bandwidth. To achieve 30X coverage, a lab would need to use three of the four lanes available in fast mode. Considering each machine costs approximately $700K, labs would need to make a significant investment to make routine testing feasible.
Finally, although the results of the Encyclopedia of DNA Elements (ENCODE) project clearly illustrate that DNA sequences outside of genes are not “junk,” determining the clinical significance of the mutations identified in these regions can be problematic.4 As a result, even though the Kingsmore group performed whole genome sequencing, they restricted their analysis to 600 recessive and mitochondrial genes to make the data clinically interpretable.
An alternative approach is to perform target enrichment for all protein coding exons in the genome (exome) and sequence only these regions. This is beneficial, as 85% of known disease-causing mutations are found in the coding region of genes or flanking splice sites, making exome sequencing an extremely efficient method for discovering the majority of allelic variants responsible for rare disorders.5 This method is also cost-effective and applicable for high-throughput as it requires only ~5% to 15% as much sequencing as whole genome. Consequently, the coverage of coding regions is extremely high compared to whole genome sequencing, which increases accuracy and reduces false positive calls.
Recent successes utilizing exome sequencing in diagnosing rare Mendelian disorders are abundant.6-11 For example, a DNA sample was recently received from a one-year-old female with clinical symptoms suggestive of Leigh syndrome, a mitochondria-associated disorder, which includes difficulty swallowing, motor delay, and involuntary eye movement. It is a devastating disease in which 50% of affected individuals die by the age of three.12 The parents were desperate to identify the gene responsible in order to confirm the diagnosis and mitigate the risk of future pregnancies. Following exome targeted sequencing, a specialized bioinformatic pipeline was able to reduce more than 10,000 initial variants to 46 candidate genes. Further medical review narrowed down the list to a single gene that coincided with the patient’s clinical phenotype.
The patient harbored two unique missense mutations in the C20orf7 gene, a nuclear encoded mitochondrial inner-membrane protein with a role in the mitochondrial complex I assembly, causing mitochondrial complex I deficiency. As expected, co-segregation analysis of the mutations revealed that each parent was a carrier of one alteration. Since mitochondrial conditions have extensive clinical overlap and there are currently more than 100 known genes linked to mitochondrial energy production disorders, the diagnosis and identification of the deleterious gene would have been nearly impossible before the introduction of NGS.13 Knowing the proper diagnosis and specific genetic mutations not only advises the physician how to treat the patient but also gives the parents the opportunity for future family planning options and in utero genetic testing with help of CVS or amniocentesis.
In the last five years, technological advancements in NGS and target enrichment methods have resulted in the identification of genes responsible for more than 40 rare disorders.14 These include numerous diseases in which the genetic basis had remained elusive, such as Miller syndrome, Kabuki syndrome, Schinzel-Giedion syndrome, and metachondromatosis.8-10,15 Although tremendous strides have been made, there is still a significant number of genetic disorders for which no disease locus is known. The Online Inheritance in Man (OMIM) database, which focuses on inherited human diseases, reports there are currently 1,766 genetic disorders with a Mendelian phenotype and no causative gene identified. As NGS becomes more commonplace and is utilized routinely, we will uncover the genetic basis of these rare disorders and provide a major paradigm shift in clinical practice in terms of diagnosis, treatment, and understanding of rare diseases.
References
- Gusella JF, Wexler NS, Conneally PM, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306(5940):234-238.
- GeneTests Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle. 1993-2012. http://www.genetests.org. Accessed October 17, 2012.
- Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4(154):DOI: 10.1126/scitranslmed.3004041.
- Stamatoyannopoulos JA. What does our genome encode? Genome Res. 2012; 22(9):1602-1611.
- Cooper DN, Krawczak M, Antonorakis SE. The nature and mechanisms of human gene mutation. In: Scriver C, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. 7th Ed. New York: McGraw-Hill; 1995:259–291.
- Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096-19101.
- Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272-276.
- Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42(1):30-35.
- Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790-793.
- Hoischen A, van Bon BW, Gilissen C, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483-485.
- Janer A, Antonicka H, Lalonde E, et al. An RMND1 mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect. Am J Hum Genet. 2012;91(4):737-743.
- Thorburn DR, Rahman S. Mitochondrial DNA-associated Leigh Syndrome and NARP. In: Pagon RA, Bird TD, Dolan CR, et al, editors. GeneReviews™ (online). Seattle (WA): University of Washington, Seattle; 1993-current. http://www.ncbi.nlm.nih.gov/books/NBK1173/. Accessed October 17, 2012.
- Kirby DM, Thorburn DR. Approaches to finding the molecular basis of mitochondrial oxidative phosphorylation disorders. Twin Res Hum Genet. 2008;11(4):395-411.
- Gilissen C, Hoischen A, Brunner HG, et al. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12(9):228.
- Sobreira NL, Cirulli ET, Avramopoulos D, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PloS Genet. 2010;6(6):e1000991.