Cell cycle deregulation influences many cancers.1 Although in the current standard of care (SOC), clinicopathological factors and events at the molecular level largely determine diagnostic, therapeutic, and patient management pathways in cancer, the patient-to-patient variability in therapy response, as well as in cancer remission and recurrence, can be further influenced by the degree of deregulation of key cell cycle events. This can potentially introduce considerable unknown variability from person to person. On top of that, these deregulation profiles on their own, as well as how the tumor microenvironment interacts with them, can potentially introduce unknown variability within a tumor and/or across different tumors in a patient. This, in turn, can manifest as intra- and/or inter-tumor heterogeneity.
For example, in breast cancers, clinicopathological factors such as tumor subtype, hormone receptor status (estrogen receptor (ER) and progesterone receptor (PR)), HER2 status, nodal status, tumor stage, patient’s age, general health, menopausal status, inherited breast cancer genes such as mutations in BRCA1 and BRCA2 decide the treatment plan.2 There are other molecular tests, such as certain multi-gene panels, that can be used additionally if they meet the appropriate conditions.2
At the level of molecular biomarkers, there are some prognostic or predictive molecular tests that have successfully incorporated some of the cell cycle deregulation components at the protein or gene levels. For example, tests that incorporate p53 gene deregulation are used in breast and bone cancers, leukemia, and sarcoma if certain other conditions based on family history, symptoms, and previous cancer diagnoses are met. In breast cancer, the multi-gene panels mentioned above and, in general, other similar genomic tests have had some success by partially incorporating cell-cycle-driven gene components among many other components, such as proliferative components throughout the cell cycle (which is correlated with the Ki67 expression profile as well), G2M checkpoints, DNA repair pathways, PI3K/AKT/mTOR pathways, on top of incorporating ER, PR, and HER2 profiles.3,4 Note that ER, PR, and HER2 profiles in a tumor are also tied to key drivers of cell cycle G1/S phase.5-7
However, a substantial opportunity exists for inter- and intra-person variability and intra- and/or inter-tumor heterogeneity to influence the landscape of therapy response, recurrence, and remission in breast cancer, and across a spectrum of other cancers.8-10 On the other hand, most of the neoadjuvant and adjuvant targeted therapies across chemotherapy, immunotherapy, and other targeted therapies (small molecules or monoclonal antibodies) used in the current standard of care across multiple cancers are agents that are designed to precisely target the cell cycle.1 For example, Table 1 refers to various commonly used neoadjuvant chemotherapy and adjuvant lines of chemotherapy in breast and other cancers. All of them precisely target certain events specific to various phases of the cell cycle. From the molecular point of view, tests measuring mutations or deficiencies in DNA damage and repair pathways such as BRCA mutational status, homologous recombination deficiency (HRD) status, etc. are natural candidates who could possibly explain sensitivity to some of these agents, such as sensitivity towards platinum-based chemotherapy in advanced ovarian cancer.11 Thus, they can help guide cohort selection.However, a more precise therapy response measurement covering the aspect of patient-to-patient and interpatient variability could potentially come from the cell cycle S, S/G2, and M phase-specific surrogate biomarkers and presumably an envelope biomarker collectively encompassing these components. Needless to say, if such cell cycle–targeted surrogate biomarkers, with or without immune interactions, are able to predict clear therapy response signatures in terms of predicting pathologic complete response (pCR) or residual cancer burden (RCB) post-neoadjuvant therapy, it would dramatically impact the efficiency of novel biomarkers in cancer and hopefully facilitate, to some extent, regulatory and payor considerations around their development endeavor.
With the advent of digitization of tissue slides, the use of digital pathology workflows and PAC (picture archiving and communication) systems has started to become more commonplace beyond radiology. This year, the American Medical Association (AMA) CPT Editorial Panel published 13 new add-on category III temporary CPT codes (0751T–0763T) for digital pathology digitization procedures, which can be used to report additional clinical staff work and service requirements associated with digitizing glass slides for primary diagnosis.12 This has created enthusiasm for adopting digital pathology workflows.
As a next step in digital pathology, a natural question can arise around how “digitally extracted” biomarkers from hematoxylin and eosin (H&E) whole slide images (WSIs) can support prognostication and therapy response prediction. In particular, how could an automated digital pathology workflow possibly support the development of cell cycle and immune interaction-targeted surrogate biomarkers generated from H&E WSIs, and how could they achieve a viable reimbursement pathway?
At present, the majority of the available digital pathology diagnostic tools, primarily based on standard artificial intelligence/machine learning (AI/ML) methods and data-driven learning combining digital pathology and clinical data, are either being developed or deployed for research use only (RUO) to identify cell-type-specific or tissue segmentation–specific diagnostic features. In the oncology space, the potential clinical application of these solutions is currently limited to immunohistochemistry (IHC) scoring and cell-type-based scoring of immune compartments.13 There is currently no reimbursement code available for these solutions. Hopefully, new digital pathology add-on codes will be available in this category in the near future.
Beyond the scope of any standard AI/ML tools, a tumor biology-driven computational digital pathology solution would be very effective if it could extract cell cycle deregulation signatures directly from the digital images of biopsy, resection, or cytology specimens, and if possible, from the routine H&E-stained specimens alone.9,14,15 The simplistic analytical ability of such a solution to quantify these deformations from the digital WSIs of H&E-stained tissues alone can bring forward novel collective information independent of routine clinicopathological factors, a key ingredient in predicting therapy response. Such collective information can be even more relevant for large or heterogeneous tumors, where therapy response is likely driven by competitions between different clonal populations and an overall knowledge of the tumor might be necessary.
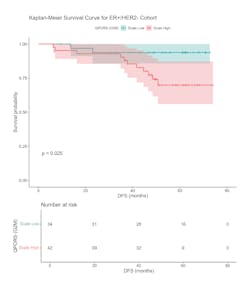
For example, using a statistical physics and tumor biology-based computational methodology, the cell-cycle G2/M deregulation-based biomarker extracted from the digital WSIs of H&E-stained ER+/HER2- breast cancer excisions was prognostic for disease-free survival for node-negative patients over a 5-year median follow-up period (p = 0.025, n = 76).14 Moreover, the biomarker could predict 20% more recurrences in a low-risk population designated by a standard of care test. All the samples in this case were drawn from pre-treatment excisions, and the biomarker primarily represented malignant cell cycle deregulations coming from cell cycle G2/M perturbations (see Figure 1).Such a solution could potentially be a very useful tool for developing pan-cancer biomarkers that are prognostic of progression-free or disease-free survival and are capable of predicting tumor recurrences. More importantly, such a solution can possibly provide biological insight to tumor recurrence and resistance to therapy for a wide variety of treatment pathways.
This type of computational digital pathology platform (with appropriate regulatory clearance) can simultaneously be used as an adjunct for reporting selected routine reflex tests (such as ER, PR, HER2, and Ki67 status in breast cancer.9,15,16 With the ability of this category of solutions to generate routine reflex tests from standard digital inputs (H&E WSIs) and deliver them in the standard reporting format following the current SOC, it is possible some existing CPT codes could be leveraged under proper allowance from appropriate authorities, such as the AMA. However, the whole new nature of these solutions, expanding to predictive and prognostic tests, might demand new CPT codes or other reimbursement pathways.
References
1. Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226(2):352-364. doi:10.1002/path.3022.
2. Breast cancer - types of treatment. Cancer.net. Published June 25, 2012. Accessed August 22, 2023. https://www.cancer.net/cancer-types/breast-cancer/types-treatment.
3. Klein ME, Dabbs DJ, Shuai Y, et al. Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Mod Pathol. 2013;26(5):658-64. doi:10.1038/modpathol.2013.36.
4. Haan JC, Bhaskaran R, Ellappalayam A, et al. MammaPrint and BluePrint comprehensively capture the cancer hallmarks in early-stage breast cancer patients. Genes Chromosomes Cancer. 2022;61(3):148-160. doi:10.1002/gcc.23014.
5. Petrossian K, Kanaya N, Lo C, et al. ERα-mediated cell cycle progression is an important requisite for CDK4/6 inhibitor response in HR+ breast cancer. Oncotarget. 2018;12;9(45):27736-27751. doi:10.18632/oncotarget.25552.
6. Koirala N, Dey N, Aske J, De P. Targeting Cell Cycle Progression in HER2+ Breast Cancer: An Emerging Treatment Opportunity. Int J Mol Sci. 2022;11;23(12):6547. doi:10.3390/ijms23126547.
7. Bower JJ, Vance LD, Psioda M, et al. Patterns of cell cycle checkpoint deregulation associated with intrinsic molecular subtypes of human breast cancer cells. NPJ Breast Cancer. 2017;31;3:9. doi:10.1038/s41523-017-0009-7.
8. Li A, Keck JM, Parmar S, et al. Characterizing advanced breast cancer heterogeneity and treatment resistance through serial biopsies and comprehensive analytics. NPJ Precis Oncol. 2021;26;5(1):28. doi:10.1038/s41698-021-00165-4.
9. Mukhopadhyay S, Dasgupta T, Orsi NM, Cummings M, Berwick A. Automated, IHC/FISH-free, H&E histopathology image-based HER2 amplification, tumor infiltrating lymphocyte (TIL) and tumor heterogeneity profiling in breast cancer. J Clin Oncol. 2021;39(15_suppl):593-593. doi:10.1200/jco.2021.39.15_suppl.593.
10. Baliu-Piqué M, Pandiella A, Ocana A. Breast Cancer Heterogeneity and Response to Novel Therapeutics. Cancers (Basel). 2020;5;12(11):3271. doi:10.3390/cancers12113271.
11. Feng Z, Shao D, Cai Y, et al. Homologous recombination deficiency status predicts response to platinum-based chemotherapy in Chinese patients with high-grade serous ovarian carcinoma. J Ovarian Res. 2023;15;16(1):53. doi:10.1186/s13048-023-01129-x.
12. 2023 digital pathology codes. College of American Pathologists. Published December 21, 2022. Accessed August 22, 2023. https://www.cap.org/advocacy/payments-for-pathology-services/2023-digital-pathology-codes.
13. Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35(1):23-32. doi:10.1038/s41379-021-00919-2.
14. Mukhopadhyay S. Prediction of disease recurrence in low risk Oncotype Dx breast cancers from digital H&E-stained whole slide images of pre-treatment resections alonee. Mukhopadhyay.
15. Dasgupta T, Mukhopadhyay S, Orsi NM, Cummings M, Berwick A. Automated immunostaining-free prediction of breast carcinoma ER and PR status, Ki67 count, patient therapy stratification index and quantification of prognostic quiescence burden in TNBC from pretreatment H&E stained histopathology images in breast cancer. J Clin Oncol. 2021;39(15_suppl):e12535-e12535. doi:10.1200/jco.2021.39.15_suppl.e12535.
16. Inacio P. 4D path’s breast cancer diagnostic platform named breakthrough device. Breast Cancer News. Published November 30, 2020. Accessed August 22, 2023. https://breastcancer-news.com/2020/11/30/fda-names-4d-path-software-for-breast-cancer-diagnostic-accuracy-breakthrough-device/.