Genetic diseases or disorders are caused due to abnormality or mutations in the genetic makeup. As of July 2024, Online Mendelian Inheritance in Man (OMIM) listed 7,528 single locus (Mendelian) diseases that are associated with 4913 genes,1 and new genetic disorders are constantly being described in medical journals.2 Around 80 percent of rare diseases are of genetic origin.3 Currently, around 300 million people live with rare diseases.3 Genetic disorders are broadly categorized into three types: single-gene defects, chromosomal abnormalities, and multifactorial conditions.4 Genetic diseases can be caused due to mutations in a single gene or multiple genes.5
Single gene disorders
· Autosomal disorders are caused by a mutation in one of the 22 autosomal chromosomal pairs. These can be autosomal dominant or autosomal recessive. Autosomal dominant disorders happen when a gene mutation or abnormality from one of the parents passes on the disorder to the child. A few examples of autosomal dominant disorders are Huntington’s disease, Marfan syndrome, and achondroplasia.6 Autosomal recessive disorders happen when a gene mutation of abnormality is passed on to the child from both parents. A few examples of autosomal recessive disorders are cystic fibrosis, Tay-Sachs disease, sickle cell disease, and thalassemia.7
· X-linked disorders occur when a mutation in X-chromosome is passed on to the child. There are at least 533 disorders due to the involvement of the genes on the X chromosome. A few examples of the x-linked chromosomes are: Red-green color blindness, hemophilia, and Duchene muscular dystrophy.8
· Y-linked disorders occur when a mutation in the Y-chromosome is passed on by a father to his son. A few examples of Y-linked chromosomes are Y chromosome infertility and some cases of Swyer syndrome.9
· Trinucleotide repeat disorders, also called triplet repeat disorders, are a set of over 30 genetic disorders caused by a mutation in which repeats of three nucleotides (trinucleotide repeats) increase in copy numbers until they cross a threshold above which they cause developmental, neurological, or neuromuscular disorders. Depending on its location, the unstable trinucleotide repeat may cause defects in a protein encoded by a gene; change the regulation of gene expression; produce a toxic RNA; or lead to production of a toxic protein.10,11,12 A few examples of trinucleotide repeat disorders are myotonic dystrophy (DM), Huntington disease, spinocerebellar ataxia, Friedreich ataxia, and fragile X syndrome.13
· Mitochondrial mutation disorders can affect one part of the body or many parts, including the brain, muscles, kidneys, heart, eyes, and ears, as mitochondria are the powerhouse of energy. Examples of mitochondria diseases are Barth syndrome, chronic progressive external ophthalmoplegia, Kearns-Sayre syndrome, Leigh syndrome, mitochondrial DNA depletion syndromes, mitochondrial encephalomyopathy, lactic acidosis, mitochondrial neurogastrointestinal encephalomyopathy, myoclonic epilepsy with ragged red fibers, neuropathy, ataxia, retinitis pigmentosa, and Pearson syndrome.14
· Imprinting disorders are congenital conditions that are due to disturbances of genomic imprinting. Genomic imprinting is the process by which only one copy of a gene in an individual (either from their mother or father) is expressed, while the other copy is suppressed. A few examples of imprinting disorders are Prader–Willi syndrome, Angelman syndrome, and Beckwith–Wiedemann syndrome.15
Chromosomal disorders
Chromosomal disorders are characterized by a morphological or numerical alteration in single or multiple chromosomes, affecting autosomes, sex chromosomes, or both.4 Chromosome abnormalities occur due to an error in cell division (mitosis or meiosis), which may occur in the prenatal, postnatal, or preimplantation periods. Chromosomal disorders are classified as numerical or structural and constitutional or acquired. A few examples of chromosomal disorder are:
· Down's syndrome or trisomy 21
· Edward's syndrome or trisomy 18
· Patau syndrome or trisomy 13
· Klinefelter's syndrome or presence of additional X chromosome in males
· Turner syndrome or presence of only a single X chromosome in females
Multifactorial disorders
Multifactorial disorders, also called complex disorders, occur due to the influence of multiple genes acting in concert with environmental factors.5 This class of disorders probably represents the single largest class of inherited disorders affecting the human population. Heart disease, diabetes, cancer, asthma, schizophrenia, osteoporosis are all examples of multifactorial disorders.
Testing for genetic diseases
Genetic testing involves analysis of DNA, chromosomes, or proteins from an individual’s blood, skin, hair, or other tissue specimens for a change, or mutation, which is associated with a genetic condition. When a mutation occurs, it may affect all or part of a gene and can result in an abnormal function leading to disease.16 Three major types of genetic testing are available in laboratories: cytogenetics examine structure and number of chromosomes under the microscope, biochemical tests measure proteins produced by genes, and molecular tests look for DNA mutations 16
Cytogenetics: This testing involves the analysis of cells in a sample of blood, tissue, amniotic fluid, bone marrow, or cerebrovascular fluid to identify any changes in an individual’s chromosomes. There are three major methods of cytogenetic testing:
Standard karyotyping: The first cytogenetics method used in the 1950s to visualize human chromosomes and their differences linked to conditions such as Down syndrome. In this method, chromosomes are paired and arranged in a standardized format known as karyotype or karyogram to provide a genome-wide snapshot of an individual’s chromosomes. Karyotypes are prepared using standardized staining procedures to reveal characteristic structural features for each chromosome. The stain most commonly used is Giemsa stain and produces G-banding at the site of the condensed chromosomes.17
Karyotyping is one of the most preferred methods to detect structural and numerical abnormalities and diagnose specific birth defects, genetic disorders, and even cancers. Clinical cytogeneticists analyze human karyotypes to detect gross genetic changes—anomalies involving several megabases or more of DNA. Karyotypes can indicate changes in chromosome number associated with aneuploid conditions, such as trisomy 21 (Down syndrome), as well as more subtle structural changes, such as chromosomal deletions, duplications, translocations, or inversions.
Fluorescent in situ hybridization (FISH): A molecular cytogegenetic technique developed in the early 1980s that uses fluorescent probes to bind to particular parts of a nucleic acid sequence (DNA or RNA) with a high degree of sequence complementarity. In this technique, the full set of chromosomes from an individual is affixed to a glass slide and then exposed to a fluorescently labeled probe. The fluorescently labeled probe finds and then binds to its matching sequence within the set of chromosomes. With the use of a fluorescent microscope, the chromosome and sub-chromosomal location where the fluorescent probe bound can be seen.18 It is utilized to diagnose genetic diseases, gene mapping, and identification of chromosomal abnormalities, and may also be used to study comparisons among the chromosomes' arrangements of genes of related species.
Comparative genomic hybridization (CGH) and array comparative genomic hybridization (aCGH): Comparative genomic hybridization (CGH) is a molecular cytogenetic method to analyze copy number variations (CNVs) without the need for culturing cells. This technique aims to quickly and efficiently compare two genomic DNA samples — a test and a reference arising from two sources — that are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions. This technique was originally developed to evaluate the differences between the chromosomal complements of solid tumor and normal tissue.19
CGH has an improved resolution of 5–10 megabases compared to giemsa banding karyotyping and fluorescence in situ hybridization (FISH) that are limited by the resolution of the microscope utilized.20, 21
Array comparative genomic hybridization (aCGH): Also called as microarray-based comparative genomic hybridization, matrix CGH, array CGH is a molecular cytogenetic technique for the detection of chromosomal copy number changes on a genome-wide and high-resolution scale.22 Array CGH compares the patient's genome against a reference genome and identifies differences between the two genomes, and thereby locates regions of genomic imbalances in the patient, utilizing the same principles of competitive fluorescence in situ hybridization as traditional CGH.
With the introduction of array CGH, the main limitation of conventional CGH, a low resolution, is overcome. In array CGH, instead of using metaphase chromosomes as in traditional CGH, cloned DNA fragments (+100–200 kb) are used of which the exact chromosomal location is known. This allows the detection of aberrations in more detail - makes it possible to map the changes directly onto the genomic sequence with much higher resolution compared to traditional CGH.23 Using this method, copy number changes at a level of 5–10 kilobases of DNA sequences can be detected. This method allows one to identify new recurrent chromosome changes such as microdeletions and duplications in human conditions such as cancer and birth defects due to chromosome aberrations.24
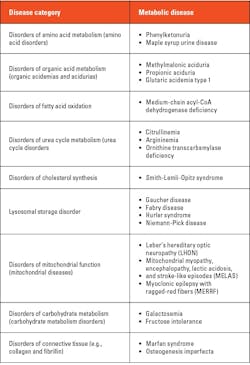
Biochemical genetic testing: Some individuals exhibit “inborn errors of metabolism,” a genetic abnormality from birth that affects their body’s metabolism. Biochemical genetic tests are highly complex tests that are used to evaluate enzyme activity; functional status of proteins; and levels of metabolites such as amino acids, organic acids, and fatty acids from a wide variety of specimen types such as urine, whole blood, plasma, serum, cerebrospinal fluid, muscle biopsy, and other tissues. These tests are used to evaluate and diagnose, monitor treatment, and clinically manage such individuals.25 Table 1 below provides a few examples of metabolic disorders for which biochemical genetic tests are performed.25
Molecular tests: These techniques have greatly advanced over the years and act as powerful tools for diagnosis, genetic consultation, and prevention of genetic diseases.26 These are also used as follows:
· Determine presymptomatic individuals' illness risk
· Detect asymptomatic recessive trait carriers
· Diagnose prenatal conditions not yet evident in pregnancy
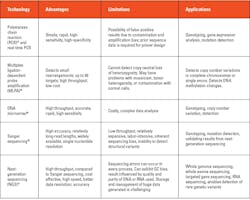
Several factors are considered when selecting the appropriate test, including suspected conditions and their possible genetic variations.26 A broad genetic test — whole genome sequencing or whole exome sequencing — is employed when a diagnosis is uncertain. A targeted test is preferred for suspected specific conditions. A targeted test could be for a single gene or a panel of genes and could be to detect point mutations, deletions, insertions, or copy number variations. Gene expression tests are performed to detect whether genes are active (producing mRNA and protein) or inactive. Too much activity (overexpression) or too little activity (underexpression) of specific genes may suggest particular genetic disorders, including various cancer types. Some of the commonly used methods used in molecular testing and their specific applications are provided in Table 2 below.
Conclusion
To date there are 7,000 types of rare diseases and 300 million people live with rare diseases.3 80% of rare diseases are due to genetic causes, which means around 240 million people live with genetic disease. Hence genetic disease is a huge burden to the world. However, due to the complexity and variability, many genetic diseases are still not diagnosed and do not have proper treatment. That said, we can be hopeful that with the advancement of science and technology that is happening at such a rapid pace we would be successful in offering precise solutions to patients with genetic diseases in the near future.
The advancements are taking place in various areas; for example, CRISPR/Cas systems and small interfering ribonucleic acid (RNA) are helping to expand our understanding of genetics at the molecular level and various omics technology along with artificial intelligence are enabling better diagnosis and monitoring of genetic diseases and cell and gene therapy, which help to better manage patients having genetic diseases.
References
1. Gene map statistics - OMIM. Omim.org. Updated August 1, 2024. Accessed August 2, 2024. https://www.omim.org/statistics/geneMap.
2. Orphanet: Quality charter About rare diseases. Orpha.net. Accessed August 2, 2024. https://www.orpha.net/en/other-information/about-rare-diseases.
3. The Lancet Global Health. The landscape for rare diseases in 2024. Lancet Glob Health. 2024;12(3):e341. doi:10.1016/s2214-109x(24)00056-1.
4. Queremel Milani DA, Tadi P. Genetics, Chromosome Abnormalities. StatPearls Publishing; 2023.
5. CDC. Genetic disorders. Genomics and Your Health. Published May 17, 2024. Accessed August 2, 2024. https://www.cdc.gov/genomics-and-health/about/genetic-disorders.html.
6. Autosomal dominant. Medlineplus.gov. Accessed August 2, 2024. https://medlineplus.gov/ency/article/002049.htm.
7. Gulani A, Weiler T. Genetics, Autosomal Recessive. StatPearls Publishing; 2023.
8. Basta M, Pandya AM. Genetics, X-Linked Inheritance. StatPearls Publishing; 2023.
9. What are the different ways a genetic condition can be inherited? Medlineplus.gov. Accessed August 2, 2024. https://medlineplus.gov/genetics/understanding/inheritance/inheritancepatterns/.
10. Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575-621. doi:10.1146/annurev.neuro.29.051605.113042.
11. Boivin M, Charlet-Berguerand N. Trinucleotide CGG Repeat Diseases: An Expanding Field of Polyglycine Proteins? Front Genet. 2022;28;13:843014. doi:10.3389/fgene.2022.843014.
12. Depienne C, Mandel JL. 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am J Hum Genet. 2021;6;108(5):764-785. doi:10.1016/j.ajhg.2021.03.011.
13. Paulson H. Repeat expansion diseases. Handb Clin Neurol. 2018;147:105-123. doi:10.1016/B978-0-444-63233-3.00009-9.
14. Mitochondrial disorders. National Institute of Neurological Disorders and Stroke. Accessed August 2, 2024. https://www.ninds.nih.gov/health-information/disorders/mitochondrial-disorders.
15. Eggermann T, Monk D, de Nanclares GP, et al. Imprinting disorders. Nat Rev Dis Primers. 2023;29;9(1):33. doi:10.1038/s41572-023-00443-4.
16. Genetic Alliance, The New York-Mid-Atlantic Consortium for Genetic and Newborn Screening Services. Genetic Testing. Genetic Alliance; 2009.
17. O’connor C. Karyotyping for chromosomal abnormalities. Nature Education. 2008;1(1).
18. Fluorescence in situ hybridization (FISH). Genome.gov. Accessed August 2, 2024. https://www.genome.gov/genetics-glossary/Fluorescence-In-Situ-Hybridization.
19. Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;30;258(5083):818-21. doi:10.1126/science.1359641.
20. Strachan T, Read A. Human Molecular Genetics. Garland Science; 2010.
21. Weiss MM, Hermsen MA, Meijer GA, et al. Comparative genomic hybridisation. Mol Pathol. 1999;52(5):243-51. doi:10.1136/mp.52.5.243.
22. Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37 Suppl:S11-7. doi:10.1038/ng1569.
23. Oostlander AE, Meijer GA, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet. 2004;66(6):488-95. doi:10.1111/j.1399-0004.2004.00322.x.
24. Ren H, Francis W, Boys A, et al. BAC-based PCR fragment microarray: high-resolution detection of chromosomal deletion and duplication breakpoints. Hum Mutat. 2005;25(5):476-82. doi:10.1002/humu.20164.
25. Centers for Disease Control and Prevention (CDC). Good laboratory practices for biochemical genetic testing and newborn screening for inherited metabolic disorders. MMWR Recomm Rep. 2012;61(RR-2):1-44.
26. Ishida C, Zubair M, Gupta V. Molecular Genetics Testing. StatPearls Publishing; 2024.
27. Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol. 2013;133(3):1-4. doi:10.1038/jid.2013.1.
28. Stuppia L, Antonucci I, Palka G, Gatta V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int J Mol Sci. 2012;13(3):3245-3276. doi:10.3390/ijms13033245.
29. Bumgarner R. Overview of DNA microarrays: types, applications, and their future. Curr Protoc Mol Biol. 2013 Jan;Chapter 22:Unit 22.1. doi:10.1002/0471142727.mb2201s101.
30. Frost A, van Campen J. Sanger sequencing. GeNotes. Accessed August 2, 2024. https://www.genomicseducation.hee.nhs.uk/genotes/knowledge-hub/sanger-sequencing/.
31. Lohmann K, Klein C. Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics. 2014;11(4):699-707. doi:10.1007/s13311-014-0288-8.
About the Author

Bipin Chandra Dash, PhD
has his doctorate in Life Science with major in Genetics and Masters in Business Administration (MBA) with major in Marketing. His areas of research are Biochemistry & Molecular Biology, Genetics, Immunology, Oncology, Virology, Tissue Culture, Vaccine R&D, etc.