The majority of molecular methods available to today’s clinical laboratory for detection of specific nucleic acid sequences employ some form of nucleic acid amplification—primarily as variations of PCR or RT-PCR, but also including techniques such as nucleic acid sequence-based amplification (NASBA, ligase-mediated chain reaction [LCR], or loop-mediated isothermal amplification (LAMP). Indeed, much of the power of molecular methods arises from the exquisite sensitivity afforded by nucleic acid amplification, whereby as little as a single appropriate target region of DNA or RNA can be selected from a complex sample and generate a specific and readily detected signal.
In addition to common strengths, nucleic acid amplification-based methods also share some common downsides. One is that they all require specific enzymes to function, and these enzymes need to work reliably within specification for valid results. With good production quality control and good logistics, this is a relatively small challenge. Approaches such as glassification or lyophilisation to stabilize enzymes against ambient temperature stress can also be employed in those situations where the logistics side is poor, such as remote fieldwork.
A more challenging weakness of amplification-based methods is that they are not intrinsically well-suited to quantitation of the target analyte; the combination of non-linear amplification (exponential, in most methods), stochastic effects, and variable presence of inhibitory substances in samples all make quantitation highly challenging. Of course, as most readers will know, robust and reliable quantitative PCR and RT-PCR methods exist and are widely used. This is purely thanks to elegant combinations of chemistry (appropriate controls, calibrator standard curves), engineering (instrument sensitivity capable of detecting signals during early amplification, before complicating factors can compound), and microfluidics/statistics/data processing (for example, digital PCR). That these solutions exist and work as well as they do is a testament to human ingenuity and resourcefulness, not a reflection of the underlying method’s intrinsic suitability for quantitative applications. In moderate to highly multiplex assays, however, even these compensatory mechanisms are not enough to ensure even, unbiased representation of all targets being quantitatively analyzed.
Non-amplified nucleic acid detection methodologies trade off a loss in sensitivity for an inherent suitability to generating quantitative data. As no (or at least, fewer) molecular manipulations are involved between the target analyte sequences and the resulting signal, more linear and thus more directly interpretable quantitative signals are produced. Depending on the precise non-amplified method employed, there can be less stringent requirements around enzymatic function in the process, leading to less dependence on the good reagent logistics chain mentioned above. Additionally, the reduced sensitivity of these methods as compared to classical PCR may in some instances be considered beneficial, as it removes the dreaded risk of carryover amplicon contamination.
All of the non-amplified molecular methods employ hybridization of sequence-specific probes to their target analyte complement(s) as initial steps. In order to have lower bound detection (sensitivity) limits of any real utility, most of these approaches rely either on a probe-based signal integration over time or on a signal amplification method. The simplest and most readily visualized of these methods hail from the early days of molecular biology: Southern and Northern blots. (Southern blotting was named after its developer, Edwin Southern; by analogy, similar conceptual approaches including Northern, Western, Southwestern, and Northwestern blotting were then named for compass directions). Southern blots involve cutting a bulk DNA sample with restriction enzymes into a reproducible mixture of small fragments, size-separating these fragments by gel electrophoresis, transferring and binding the size-separated fragments to a solid support such as nylon, and then hybridizing a labeled DNA probe of analyte-complementary sequence across the support surface. Excess unhybridized probe is removed by washing, and bound probe is visualized via its label—originally, a radioisotope. Exposure of the blot to film allowed for a visual identification of analyte positivity, with sensitivity controlled by length of exposure time (as much as weeks for very faint signals). Use of phosphorimager devices instead of film allowed for direct quantitative measurement of signal intensities, which could readily be interpreted against standards to quantitate analyte. Northern blotting is exactly analogous, but performed on RNA targets.
While still useful in research, both of these methods have mostly been supplanted in modern clinical settings by microarray hybridization approaches (see The Primer, “Applications of multiplexing and array method,” MLO, September 2013). The probe label of choice is now generally a fluorophore as opposed to a radioisotope, but sensitivity is still achieved through (optical) signal integration times.
A related approach which also bypasses the need for nucleic acid amplification is FISH; as this was the subject of a recent (August 2015) installment of this series, we won’t cover it in further detail here.
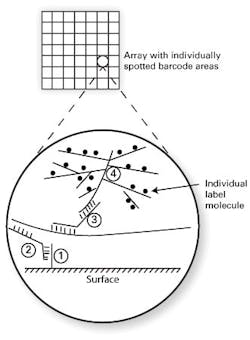
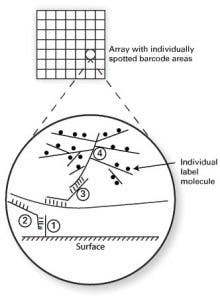
Two methods which bypass nucleic acid amplification bear particular mention because of their relatively frequent occurrence in clinical laboratory settings. The first of these is bDNA, or branched chain DNA. As the name implies, this works through the synthetic creation of highly branched, “tree-like” structures of single-stranded DNA. One format of this assay type uses a solid assay surface, coated in spots (arrays) with particular DNA “barcodes” or address sequences (see Figure 1, DNA strand “1”). A second piece of single-stranded DNA (“2”) is designed to hybridize on one end to this barcode, with its other end hybridizing to the target analyte sequence. If the target analyte is present, this “sandwich” or “bridge” hybridization now tethers the target analyte to a specific array location. A third single-stranded DNA probe (“3”) is included, which also shares half complementarity to the analyte (thereby binding to the localized, bound analyte) and on its other end, binds a highly branched, tree-like structure which has many labeled arms as its “branches” (“4”). This order (base/ bridge from base to specific target/ bridge from specific target to branched label/ branched label) thus effectively puts very large numbers of labels such as fluorophores on the array spot barcoded to the target of interest.
The degree of branching on the label can support up to thousands of individual label molecules, making a bright enough signal to directly detect the very small numbers of analyte molecules. The purely hybridization-based approach, without nucleic acid amplification, makes the signal intensity much more reliably linear with regard to target material than amplified methods. This approach simultaneously delivers good sensitivity, good ability to quantify analyte, lower susceptibility to contamination than amplified methods, and high multiplexing capacity. These strengths have led to there being a range of clinical assays available in bDNA format, in use for applications such as viral load monitoring or in measurement of RNA levels in formalin fixed, paraffin embedded (FFPE) tissue.1
The second method is through the use of colloidal gold nanoparticles as hybridization probe labels. In effect, the colloidal gold particle replaces the fluorophore from any other DNA or RNA probe as a 5′ or 3′ end tag. When prepared appropriately and of the right size range, these gold particles exhibit strong optical effects through an effect known as surface plasmon resonance (SPR). While the physics behind SPR is beyond the scope of this article, two facts are particularly relevant. One is that the effect creates a narrow color band light scattering, in effect “shining” with an apparent color when illuminated, and this “shining” is extremely bright—on the order of hundreds of thousands of times brighter than a single fluorophore. The second is that the apparent color is controlled by the gold nanoparticle size, such that it is in effect tunable and allows for multiplexing of different apparent colors in an assay. (This may sound familiar to readers who have come across quantum dots, which are highly efficient fluorophores with a cage-like organometallic structure, tunable for emission wavelength by varying the cage size. While similar, quantum dots are true fluorophores and not as bright on a per-label basis as SPR labels.)
The light-scattering effect of the gold nanoparticles can be further chemically enhanced or amplified in a (quasi) linear fashion by a post-hybridization silver ion enhancement step. Like the bDNA system, this large increase in the intensity or signal-to-noise ratio of this form of label over the single (or few) fluorophore-per-target molecule approaches of classical real-time PCR or array methods allows for detection of much lower numbers of unamplified analyte molecules. Some commercial systems, FDA-cleared assays for applications such as sepsis testing, are based on this technology and are seeing increasing clinical use. In addition to lowered susceptibility to template (amplicon) contamination, the lack of a nucleic acid amplification step in these assays reduces sample-to-answer time and susceptibility to PCR inhibitory substances in the samples, making such assays popular in laboratory settings where assay speed and robustness in face of potentially variable sample quality are critical factors.
While PCR and RT-PCR thus remain the mainstays of molecular diagnostic approaches, approaches such as these which omit nucleic acid amplification strategies offer additional tools in the laboratory toolbox with features and utility ideally suited to some applications.
Reference
- Knudsen BS, Allen AN. McLerran DF, et al. Evaluation of the branched-chain DNA assay for measurement of RNA in formalin-fixed tissues. J Mol Diagn.2008;10(2):169-176.