In last month’s installment of the Primer, we touched on transcription-mediated amplification (TMA) as an alternative to polymerase chain reaction (PCR)-based strategies for the amplification and detection of specific nucleic acid sequences. We’ll carry on in that vein this month with an examination of two other methodologies used in some clinical settings: the ligase chain reaction (LCR) and helicase dependant amplification (HDA).
Ligase chain reaction
LCR was initially described very soon after the advent of PCR, and was already well established by the mid-1990s. The method is particularly useful for the detection of pre-identified single nucleotide polymorphisms (SNPs), such as those point mutations resulting in common genetic diseases. Examples of these most familiar to the reader probably include hemochromatosis (H63D, C282Y) and sickle-cell anemia (E6V). (For those readers who haven’t brushed up on their genetics notation for a while, missense mutations, where one amino acid in a protein is replaced with another, are designated by the one-letter code for the normal “wild-type” amino acid, a number representing the position of the amino acid in the polypeptide sequence, and the one letter code of the amino acid which occurs in this mutation. Frequently, these mutations arise as SNPs in the underlying DNA element coding for the protein.)
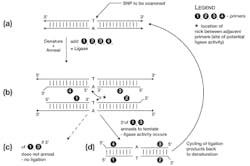
Like PCR, LCR relies on short synthetic oligonucleotides, complementary in sequence to the area of interest. In this methodology, rather than broadly flanking a region of interest, the primers lie immediately on top of the putative SNP. Two primers are complimentary to one target DNA strand, and when hybridized to template lie directly adjacent to each other, with the SNP falling exactly under the 3′ end of the upstream primer “1” (Figure 1b). If (but only if) the 3′ end of primer 1 anneals to the template, an enzyme known as DNA ligase catalyzes the covalent joining of the 3′-OH of primer 1 to the 5′ phosphate of primer 2. This results in a new DNA molecule of (primer 1 + primer 2).
Three things to note particularly here: first, that this potential (primer 1 + primer 2) product creates a copy of the opposite template strand flanking the potential SNP; second, if the 3′ nucleotide of primer 1 does not anneal to the template, ligation does not occur (at least, not at any significant rate); and third, the 3′ end of primer 2 ends in a sequence chosen specifically to not anneal to the template; this is to avoid this 3′ end being involved in any additional, unwanted ligation reactions. (A more reliable but slightly more expensive method of doing this would be to synthesize primer 2 with its 3′ nucleotide as a dideoxy nucleotide, in effect making it a DNA chain terminator as in Sanger sequencing).
This same process of annealing a primer pair and template sequence-dependant ligation of them to form a product occurs simultaneously on the other template strand, using primers 3 and 4. The same points of note apply to this primer set as did to 1 and 2.
If the template SNP matches (anneals) to the 3′ ends of primers 1 and 3, as in Figure 1c, ligation products (1+2) and (3+4) can now each be denatured, and may now act as template for the annealing and ligation of the opposite primer pair. That is, primers 3 and 4 anneal to and ligate across the (1+2) product, and primers 1 and 2 anneal to and ligate across the (3+4) product. The reaction proceeds by repeatedly cycling the reaction temperature between denaturation (95°-99°C) and a lower temperature suitable for sequence specific annealing and DNA ligase activity (depending on sequence composition and length of the primers, but generally in the 50°-60°C range). If the primers match the SNP, this results in an exponential amplification of the levels of the (1+2) and (3+4) product; if the SNP does not match, the four test primers remain unjoined throughout the cycling (Figure 1d). Analysis of the reaction products for the (1+2 / 3+4) product is then done as a means to determine if the expected SNP was present in sample, or not. This product detection can be done by any of several means covered in prior installments of the Primer, including surface or bead-array based capture techniques with fluorescently labelled products.
The astute reader will note that above, it was indicated that if the primers don’t match the SNP, ligation does not occur “at any significant rate.” Strictly speaking, ligation can still occur at very low rates when rare events known as “tautomeric shifts” in either the SNP nucleotide bases, or those at the 3′ ends of primers 1 and 3, occur. These rare electron rearrangements can allow “non-canonical” base pairing to occur very briefly, and if this happens when ligase is present, a product can still be formed. This would lead to a false positive signal in the LCR reaction, as this one primer ligation product is now capable of amplifying normally.
While the risk of this occurring is very low, there are variations on the LCR method which can reduce the risk even further. Most common of these is “gapped” LCR, which works by shortening each of primers 1 and 3 by a single, 3′ nucleotide (that is, the nucleotide testing for SNP annealing). In addition to DNA ligase, the reaction mix needs also to contain DNA polymerase and only the “missing” dNTPs. The concept here is that if the expected SNP is present, the appropriate “missing” 3′ dNTP from primers 1 and 3 will be added on by normal activity of the DNA polymerase, after which the classical LCR steps can occur. If, however, the template SNP location is not the expected nucleotide, the DNA polymerase is unable to extend primers 1 and 3 (the matching dNTP is not present). This approach adds a second layer of sequence specificity to the LCR process and can reduce false positive rates to vanishingly small values.
Helicase-dependent amplification
We start our examination of this month’s second method by considering something that PCR and TMA methods have in common, previously described. Both employ a pair of template-specific complementary primers to define the ends of the sequence to be amplified; these primers act to recruit and initiate polymerase function in making new product strands. In order for these specific primers to anneal, the template material must be single-stranded (ssDNA). In the case of PCR, thermal denaturation in each cycle creates the single-strand templates; in TMA, the initial target is single-stranded RNA and within the cycling process, RNase H activity degrades the RNA component of DNA/RNA hybrid molecules to leave ssDNA.
Within the context of a living cell, DNA replication by polymerases also initiates from primer sequences, but without either of these methods. Instead, in vivo DNA replication methods for most organisms rely on a class of enzymes known as DNA helicases, which act to locally unwind dsDNA to create small ssDNA regions suitable for polymerase activity. They are assisted in doing so by a handful of other proteins, including single-stranded DNA binding proteins (SSBs) which coat ssDNA regions to help keep them from immediately re-annealing. The method known as helicase dependant amplification (HDA) applies this approach to create an in vitro, isothermal target-specific nucleic acid amplification technology (Figure 2).
In this approach, template DNA is mixed with two template-specific primers which are of opposite orientation and flank the region of interest (just as in PCR), along with a DNA helicase, some SSBs, and often (depending on the specific helicase and polymerase), a few other “accessory proteins” which assist the helicase and DNA polymerase in optimum function. The DNA helicase acts to unwind the reaction template DNAs, and the SSBs help to keep it single-stranded long enough for the primer oligonucleotides to “search” the ssDNA regions for their potential complemetary annealing sites (Figure 2b). If these exist, the high concentration of primers aids in their displacing the SSBs and creating an annealed primer-template substrate which now recruits in the DNA polymerase. Polymerase activity extends the primer, creating a new nascent strand and, in the process, creating a new annealing site for the opposing primer (Figure 2c).
While this is occurring, DNA helicase, SSBs, and (depending on the precise reaction mixture) some other accessory factors come in and unwind the new nascent strand from the original template. (See Figure 2d, focusing just on the lower strand from 2c.) This makes the nascent strand ssDNA, allowing the opposing primer (Primer 1 in Figure 2c) to anneal, and DNA polymerase to again start strand synthesis. This time, the template strand only exists as far as the 3′ end of the opposing primer (Primer 1, Figure 2c). Repeating cycles of helicase denaturation, primer annealing, and polymerase extension act very much like classical PCR at this point, resulting in large-scale production of a primer-to-primer DNA product (Figure 2e) in those reactions where a template matching the primers existed. Unlike PCR however, this entire reaction process occurs at a single temperature (isothermal)—so while HDA requires a few more enzymes in the “master mix” than conventional PCR, it doesn’t require a complex, computer-controlled thermal cycler. Any simple water bath or other stable temperature heat block, or other method for keeping the reaction at a stable temperature, will suffice.
In practice, HDA doesn’t produce as much target amplification as a well optimized PCR reaction. However, in a one-hour reaction, it can still yield 107 or greater amplification factor. For many applications, particularly in infectious agent-detection settings, this is more than enough sensitivity, and HDA reactions capable of detecting a few hundred target copies per reaction are quite possible.
Detection of the HDA product can be done by any of the means used to detect PCR products, including endpoint analysis (for example, by gels or arrays) or by fluorescent based real-time methods. In keeping with the instrumental simplicity of HDA, however, most currently available commercial diagnostics based on this technique employ a simple variation on the instrumentation-free, visually readable “line probe assay” approach (to be covered in next month’s column on alternate detection strategies). This combination of a very simple but powerful and specific isothermal amplification strategy and a simple, disposable, instrumentation-free detection system brings a molecular diagnostic tool functionally closer to point of care (POC) testing than ever before—even if it is still constrained from that designation at present for regulatory reasons.
While last month’s and this month’s columns together have not covered all of the non-PCR nucleic acid amplification strategies in use today (loop-mediated isothermal amplification [LAMP], strand displacement amplification [SDA], and rolling circle replication [RCR] are other methods one may encounter), hopefully they will have opened the reader’s eyes to the existence of a range of competing technologies for molecular diagnostic applications. In specific instances, some of these have distinct advantages, and readers may expect to encounter some implementation of one of these methodologies in their laboratories, either now or in the future.