Most of the molecular methods covered so far in this series have involved methods for the detection of known sequences of DNA or RNA. This begs the question of how these sequences (of pathogen genes, human chromosomal mutations, and the like) were identified and characterized in the first place. Similarly, it’s clear these methods aren’t of use if you have an unknown target for which you can’t make a primer sequence (an emerging pathogen, or a novel mutation, for example). The answers to these questions hinge on techniques for DNA sequencing.
Sequencing is the name applied to any of a range of technologies which can take a DNA strand and tell us, in linear order, the “sequence” of the four possible nucleotide bases A, G, C, and T (thus the name). The data obtained from these approaches can be compared against other sequences, such as from the same organism (useful for identifying mutations or individual variations), or from related species (allowing assessment of how closely related two samples are). It is from this detailed sequence information that we are then able to design specific PCR primers or other sequence-specific targeted approaches to rapidly test large numbers of specimens to see whether they match our sequenced reference material. Sequencing techniques—while not always at the forefront of diagnostic methods—underlie almost all of the widely differing MDx technologies at some level. As such the topic is worth considering in this series, but due to the breadth and complexity, we will unfortunately be limited to only a very brief overview of the methods and applications. In particular, the next-generation sequencing (NGS) methods are technically involved, and only the most cursory descriptions are provided here.
Earliest methods of sequencing short sections of purified DNA were chemical in nature and relied on the ability to selectively break DNA strands at specific bases (G, A and G, C, and C and T) in each of four parallel reactions. By allowing reactions only to proceed partially, each reaction on a population of identical DNA template molecules left a population with some members broken at each sensitive base. Size separation by a form of gel electrophoresis allowed the researcher to then work out the original sequence from the pattern of fragment lengths. The process was laborious, awkward, and only good for short sections of DNA (read lengths).
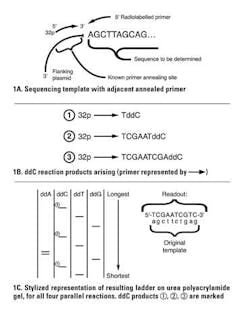
This was largely supplanted by a sequence-by-synthesis approach known as Sanger sequencing. This method also starts with a uniform population of DNA material to sequence (template). A radiolabeled primer (much like a PCR primer) is annealed down at one end of the sequence, and a DNA polymerase is then used to synthesize a new complementary strand to the target material.
You may be wondering: if a primer is needed to anneal, how can this be done for an unknown sequence? The answer is by lab manipulation of the unknown target sequence to attach it to a known DNA sequence, usually in the form of a bacterial plasmid DNA by cloning; then a single primer annealing to the adjacent known plasmid can be used to initiate synthesis down any cloned, unknown portion. The clever part in this approach is to incorporate a small percentage of “dideoxy” versions of one of the nucleotides in each of four parallel reactions. Dideoxy means that both the 2′-OH and the 3′-OH (critical as the attachment point for the next nucleotide) are missing from the nucleotide.
Consider a reaction where a small fraction of the “C” molecules are dideoxy-CTP (ddCTP). When one of these is incorporated in place of a regular dCTP, the growing nascent DNA chain terminates, as there is no 3′-OH for the polymerase to extend from. This creates a population of labeled molecules, all ending with C positions throughout the length of the template. The parallel reactions are one each with ddATP, ddGTP, and ddTTP. Products of the four reactions are electrophoresed on four adjacent lanes of a urea polyacrylamide gel (conceptually similar to an agarose gel, but with the ability to size-resolve single-base differences in DNA). Autoradiography of the gel by exposure to X-ray film allows the radiolabeled terminated DNA molecules to be visualized as a readable “ladder” of the entire sequence (Figure 1). Note that the template’s complement is what is actually read out, as shown in the figure.
An improvement to this method came when each of the ddNTPs would be labeled with a specific fluorophore—let’s say green for A, black for G, red for T, and blue for C. This meant the primer didn’t require a radiolabel, since each chain terminator simultaneously labeled the product; it also meant four reactions and lanes were not needed, as all reactions could be done together and run in a single lane, with the order of colors seen running down the gel giving the sequence. This was further improved by moving from polyacrylamide gels to small tube capillaries with a polymer matrix through which the reaction product is electophoresed. One needs simply to observe the sequence of fluorescent colors as they reach the bottom of the capillary to read out the underlying sequence from shortest (nearest the primer) to longest. Still widely in use today, capillary sequencers are fast, cheap, and easy to use for short target sequences (a few hundred to thousands of base pairs) and generate output which is instantly readable (Figure 2).
The sequence-by-synthesis approach has since been used as the basis for several NGS approaches. In one such method, a sample to be sequenced is randomly broken into short fragments which are then dispersed into tiny droplets emulsed in oil—each in theory containing a single template fragment—and deposited into microscopic individual reaction wells on a surface. The individually segregated template molecules are then observed through a real time microscope system as each of the four dNTPs is sequentially added to, and then rinsed from, the tiny reactions. In each reaction, when the appropriate next dNTP for a growing strand becomes available, it is incorporated—causing a release of the outer two phosphates (pyrophosphate) from the dNTP. In the right chemical environment, this can cause a release of light that can be captured by the microscope. An attached computer system records, for each micro reaction, which of the four dNTPs in turn reacts each round, and thus reads out the sequence in each parallel reaction. Note here that the reactions are each on different short sequences, but due to the random nature of the original DNA breakage in preparation of the micro reactions, there will be sequence overlap. The computer detects these overlaps, and “tiles” many short reads together to generate longer sequences, such as:
The massively parallel nature of this approach means that every nucleotide in a sample is read in many reactions independently (leading to high accuracy, as misread errors are rare and statistically overwhelmed by correct reads) and, in essence, an entire genome can be sampled at once.
Rather than detecting a dNTP addition by light emission as previously mentioned, it also can be detected by miniscule pH shifts in each microreaction—the basis for a similar NGS approach known as Ion Torrent. Again, a massively parallel approach producing sequences of many short reactions is used for tiling together to form longer reads.
Another NGS sequence-by-synthesis approach is paired-end sequencing. Here again, randomly sheared short template fragments are generated, then attached to known sequence ”adapters” at either end. These adapters are then used both to capture the fragments on microscopic flow cell chambers by hybridization, and as annealing points for primers to allow polymerase extension over the unknown fragment. Microscopic camera imaging is also used here, as is fluorescent chain termination at specific bases as in the Sanger capillary methods; but unlike the use of ddNTPs, this method relies on a reversible chemical chain termination. After a termination is detected, the terminator is chemically removed and the chain allowed to grow one more base, again reporting which base terminates. Parallel reactions and tiling complete the process.
Yet another NGS method, polony sequencing, starts with random shearing of target DNA, addition of known adapters to the ends of sheared molecules, and dispersion of the molecules into large numbers of (mostly) single molecule-containing microdroplets in an oil emulsion. Rather than synthesis, however, this approach now exposes each microreaction to a hybridization reaction with a short oligo containing partially a mixed sequence (representing all possible sequences) and a known fixed nucleotide, in each of the four possibilities, each labeled with a specific fluorophore. By observing which fluorophore hybridizes best, the identity of that position in the unknown sequence is obtained (it’s the complement to the best hybridizing marker). The hybridized material is removed, and the process repeated with the known base position shifted by one nucleotide. In this manner, short reads of each microreaction are again generated. By now the reader will guess this is done in a massively parallel fashion, followed by tiling.
Most NGS methods in use today use some form of one of the previously mentioned approaches, and allow for sequencing of individual genomes on a scale and speed hardly imaginable a decade ago. It is now possible to sequence the entire exome (directly coding portion) of a human sample in days for a cost below $1,000. The difficulty now comes in the computational challenges of processing, storing, and meaningfully analyzing all of this data. Particularly, it has become increasingly evident that large numbers of genetic variations can occur from one person to another; assigning clinical significance to any one of these changes can often be impossible until sufficient numbers of individuals bearing the same change, either with or without a common clinical presentation, allow correlation.
While these NGS methods are vastly improved in speed and throughput from the earliest sequencing approaches, still more leaps in cost, speed, accuracy, and throughput for whole genome sequencing may be possible with technologies now on the horizon. Particularly, “nanopore” methods which effectively take long DNA strands and “spool” them through microscopic pores—able to, in effect, feel the identity of each base in sequence as it passes through the pore—are in late-stage development. If these live up to their potential, they may be able to read much longer individual strands and require less parallelism and tiling, and afford much higher speeds than current NGS methods.
Sequencing approaches are increasingly being used clinically. Applications range from small region capillary sequencing, when a particular gene is of interest (such as a presentation suggestive of a mutation in a known gene), to NGS methods applied on tumor samples to capture a range of information helpful in identifying tumor type and viable treatment options. It is now possible to sequence an entire patient exome, or even whole genome, in relatively little time and at low cost. This is likely to become faster and cheaper, and as it does so, we may reach the point where individualized MDx tests for specific pathogens or markers become supplanted by simply determining the sequence of all genetic material in a patient sample as a routine matter. The bioinformatics, data storage, and potential ethical considerations around this are significant and will require wisdom, foresight, and understanding of the methods to ensure the best use of these technologies to improve patient care.
About the Author

John Brunstein, PhD
is a member of the MLO Editorial Advisory Board. He serves as President and Chief Science Officer for British Columbia-based PathoID, Inc., which provides consulting for development and validation of molecular assays.