Optimizing Clostridioides difficile diagnosis: Reducing overdiagnosis and enhancing antimicrobial stewardship through a two-step testing approach
Clostridioides difficile (C. diff), a Gram-positive, spore-forming bacteria, is the most common cause of hospital-acquired infection in the United States, responsible for an estimated 12,000 deaths annually.1 Clinical presentation of C. diff infection (CDI) can vary from mild cramping and diarrhea to severe colitis, with complications including pseudomembranous colitis, toxic megacolon, and septic shock. The clinical course of CDI can be recurrent, and managing this infection is complicated by antibiotic resistance and the persistence of C. diff spores in healthcare environments.
The challenge
Diagnosis of CDI is based primarily on clinical presentation and confirmed by diagnostic testing. This is complicated in an inpatient setting because non-infectious diarrhea is common in this patient population. Approximately 10% of these patients are asymptomatic carriers of C. diff,2 meaning their gut flora contains C. diff but does not exhibit clinical symptoms. Molecular testing methods for CDI such as nucleic acid amplification tests (NAAT) and polymerase chain reaction (PCR) are common and offer high sensitivity as a screening test, but they are unable to differentiate between asymptomatic carriers and patients with active CDI caused by toxin-producing C. diff organisms. As a result, C. diff testing by molecular methods alone has been shown to result in overdiagnosis and overtreatment with antimicrobial therapy.3 Because many people are asymptomatic carriers of C. diff, the Infectious Disease Society of America (IDSA), the Society for Healthcare and Epidemiology of America (SHEA), the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), and the American College of Gastroenterology (ACG) all suggest using a toxin enzyme immunoassay test to help differentiate patients with active disease from those who are simply carriers.
Additionally, a proposed change to the National Healthcare Safety Network’s (NHSN) C. diff infection reporting guidelines was announced in 2023. Previously, the last test entered into a patient record defined a CDI event. This change is intended to improve upon the existing NHSN CDI measure by including evidence of a C. diff positive test AND antibiotic treatment and defines a healthcare facility-onset, antibiotic-treated C. difficile infection (HT-CDI) as any positive C. diff test on day ≥4 and ≥5 days of C. diff antibiotic treatment.4 This proposed change is significant because the order of testing in the CDI testing algorithm no longer impacts whether a positive C. diff test is a reportable CDI event, allowing laboratories more flexibility in selecting the best algorithm for their specific patient population and workflow. With the addition of a treatment component to the proposed NHSN reporting guidelines, clinicians are also encouraged to consider clinical presentation as well as laboratory test results when diagnosing CDI.
The solution
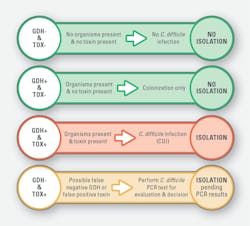
To properly determine if a patient has active CDI, the American College of Gastroenterology (ACG)5 and the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA)6 guidelines recommend a multistep testing algorithm including a sensitive screening test and a specific toxin test. Glutamate dehydrogenase (GDH) is an enzyme present in all C. diff organisms that multiple independent studies7-10 have shown to be equivalent to molecular testing methods in sensitivity for ruling out CDI. Toxin enzyme immunoassay (EIA) methods provide excellent clinical sensitivity to confirm a diagnosis of CDI. An algorithm that begins with a GDH or molecular screening test, followed by a toxin test for confirmation, has been recommended as the best performing algorithm for CDI testing.6,11
In December 2020, St. John’s Hospital, part of Hospital Sisters Health System (HSHS), transitioned from molecular screening testing only for C. diff to a complete stand-alone two-step C. diff algorithm, in a single enzyme immunoassay (EIA) test device. This change was driven by the need to comply with updated NHSN standards, aiming to lower standardized infection ratios (SIR) by improving diagnostic accuracy and reducing unnecessary isolation of colonized but uninfected patients. Additionally, the change sought to streamline infection prevention resources, as the previous method required extensive review of all orders before testing, placing a heavy burden on infection prevention teams.
The decision-making process involved key stakeholders, including the laboratory, quality improvement team, nursing, and the chief medical officer. Over a three-month period, the transition was carefully planned, encompassing validation and correlation studies, policy development, and education for both nursing and laboratory teams. Additionally, clinicians were provided with guidance on interpretation of results (See Figure 1).
The results
Following the implementation of a two-step algorithm, there was notable reduction in CDI, with a 21% decrease from 2020 to 2021, and a further 31% drop from 2021 to 2022. Overall, this led to a 45% reduction in CDI over the two-year period. This achievement reduced unnecessary patient isolations, enhanced antibiotic stewardship, and resulted in an estimated annual savings of $65,000 for the laboratory. The dual benefit of improving patient care while reducing costs aligns with HSHS’s commitment to providing high-quality, affordable healthcare in the communities they serve.
References
- Centers for Disease Control and Prevention (U.S.). Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention (U.S.); 2019.
- Miles-Jay A, Snitkin ES, Lin MY, et al. Longitudinal genomic surveillance of carriage and transmission of Clostridioides difficile in an intensive care unit. Nat Med. 2023;29(10):2526-2534. doi:10.1038/s41591-023-02549-4.
- Peng Z, Ling L, Stratton CW, et al. Advances in the diagnosis and treatment of Clostridium difficile infections. Emerg Microbes Infect. 2018;7(1):15. doi:10.1038/s41426-017-0019-4.
- Centers for Disease Control and Prevention (CDC). Introducing NHSN's new digital quality measures. https://www.cdc.gov/nhsn/pdfs/training/D1_Introducing-NHSNs-New-Digital-Quality-Measures_508c.pdf. Published June 2023. Accessed October 10, 2024.
- Kelly CR, Fischer M, Allegretti JR, et al. ACG clinical guidelines: Prevention, diagnosis, and treatment of Clostridioides difficile infections. Am J Gastroenterol. 2021;116(6):1124-1147. doi:10.14309/ajg.0000000000001278.
- McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66(7):e1-e48. doi:10.1093/cid/cix1085.
- Quinn CD, Sefers SE, Babiker W, et al. C. Diff Quik Chek Complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol. 2010;48(2):603-605. doi:10.1128/JCM.01614-09.
- Swindells J, Brenwald N, Reading N, Oppenheim B. Evaluation of diagnostic tests for Clostridium difficile infection. J Clin Microbiol. 2010;48(2):606-608. doi:10.1128/JCM.01579-09.
- Sharp SE, Ruden LO, Pohl JC, Hatcher PA, Jayne LM, Ivie WM. Evaluation of the C. Diff Quik Chek Complete Assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of Clostridium difficile disease. J Clin Microbiol. 2010;48(6):2082-2086. doi:10.1128/JCM.00129-10.
- Dubberke ER, Han Z, Bobo L, et al. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol. 2011;49(8):2887-2893. doi:10.1128/JCM.00891-11.
- Planche TD, Davies KA, Coen PG, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13(11):936-945. doi:10.1016/S1473-3099(13)70200-7.
About the Author

Jimmy Lowery MBA, MLS(ASCP)SBBCM
serves as Product Manager for TECHLAB. He is an experienced Clinical Laboratory Scientist and Product Marketer with over 20 years of experience in laboratory medicine. He has been actively involved with professional organizations like the AABB and has authored or co-authored multiple publications and posters related to laboratory testing.

Justin Cox, MBA, MLS
is the System Laboratory Director for Hospital Sisters Health System (HSHS), overseeing 13 hospital-based laboratories across the organization. With 18 years of experience, he has driven initiatives in process improvement and laboratory standardization. Justin holds an MBA from Greenville University and is recognized for his leadership and community involvement.