The history of antimicrobial resistance and the important role diagnostics plays to combat it
To take the test online go HERE. For more information, visit the Continuing Education tab.
LEARNING OBJECTIVES
Upon completion of this article, the reader will be able to:
1. Discuss the history of the discoveries and warnings of antibiotic use.
2. List the agencies and organizations involved in the development of antibiotic stewardship programs.
3. Identify the top microorganisms and antibiotics that contribute to AMR.
4. Discuss the advances in antimicrobial laboratory testing and therapies and how they contribute to combatting AMR infections.
Louis Pasteur likely predicted the concept of antimicrobial resistance (AMR) with his famous quote in the late 1800s, “Messieurs, c'est les microbes qui auront le dernier mot." (Gentlemen, it is the microbes who will have the last word.)”1
Later, penicillin was discovered on September 28, 1928, by Alexander Fleming and approved for clinical use in 1941. During his 1945 Nobel Lecture, Flemming warned of resistance through the dangers of underdosing. “It is not difficult to make microbes resistant to penicillin in the laboratory by exposing them to concentrations not sufficient to kill them, and the same thing has occasionally happened in the body. The time may come when penicillin can be bought by anyone in the shops. Then there is the danger that the ignorant man may easily underdose himself and by exposing his microbes to non-lethal quantities of the drug make them resistant.”2
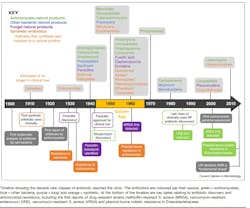
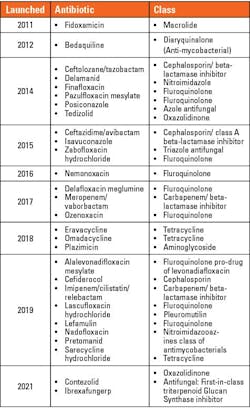
Throughout the next several decades, many classes of antibiotics were developed.3 (See Figure 1.) Interestingly, resistance to SalvarsanTM (the first clinically used antibiotic ~1910) took approximately 20 years to emerge.3,4 Resistance to sulfonamides and penicillin occurred much sooner [~12 years], as outlined in the timeline below. 3 Newer antibiotics approved since 2010 are listed in Table 1 and consist of combinations of current antibiotic classes or congeners of existing molecules.
Early in my training as a clinical pharmacist interested in infectious diseases, I can remember two specific examples of aggressive efforts by sales representatives to position an antibiotic as a preferred agent in the treatment of almost any type of infection. Such outlandish claims provide a perfect example of behaviors and perceptions within healthcare practice that contribute to the inappropriate use and overprescribing of antibiotics. These situations included a sales representative stating, “If you don’t know the bug, my new 3rd generation cephalosporin is the drug.”
At this time in the early 1990s, the sales representative knew that it could take at least 48–72 hours to get a blood or other culture result to begin to show sufficient pathogen growth to cross the threshold needed for identification and susceptibility testing (ID/AST). The sales representatives were likely trying to capitalize on the use of empiric broad-spectrum therapy to cover a wide variety of bacterial pathogens until ID/AST results became available, which could influence the use of their product.
In another memorable situation, I had a sales representative say to me “Don’t forget about my broad-spectrum fluoroquinolone for cold and flu season!” When I asked this salesperson if they realized they were potentially making an off-label claim that an antibiotic class prescription was not indicated for viral infections, he seemed offended.
These examples, while they initially may seem humorous, serve as strong representative examples of the inappropriate use of antimicrobial therapy.
As awareness and concern for antibiotic use grew, several governmental and professional organizations began raising awareness over the concern for the rising rates of resistant pathogens. These organizations leveraged the President’s Council of Advisors on Science and Technology (PCAST) to educate and enlighten government officials on antimicrobial resistance.5
In 2007 the Infectious Diseases Society of America (IDSA) published guidelines on developing institutional programs to enhance the adoption of antimicrobial stewardship practices.6 Their goal was to establish realistic guidelines for antibiotic use, due to the rising rates of antimicrobial resistance and a shrinking pipeline of de novo antibiotic approvals.
In 2013 the Centers for Disease Control and Prevention (CDC) published a report to raise awareness and establish priorities concerning resistant pathogens entitled Antibiotic Resistance Threats in the United States.7 This report set off a series of government actions in the fight against AMR (see Figure 2).8
In 2014, PCAST reported on combating antibiotic resistance to the President. An outcome of the PCAST report was the agreed goal to develop a working group of experts in antibiotic resistance in both human and veterinary sectors and to develop recommendations for the U.S. government to take actionable steps to curb the growing problem of AMR.
A Presidential executive order (13676: “Combating Antibiotic-Resistant Bacteria) was issued that accompanied the PCAST report in September 2014.9 The executive order set off a chain of initiatives, such as developing the Presidential Advisory Council on Combating Antibiotic-Resistant Bacteria (PACCARB), to forge ahead in the fight against antimicrobial resistance (Figure 2).
In 2016 the IDSA and SHEA societies published guideline updates detailing how to effectively implement an antimicrobial stewardship program.10 By the end of 2017, the Centers for Medicare & Medicaid Services (CMS) was to have federal regulations requiring the development and implementation of “robust antibiotic stewardship programs” in hospitals, critical access hospitals, and long-term care, and nursing home facilities. Outpatient antibiotic use is stated to account for ~80% of antibiotic use,11 thus it was essential for outpatient antibiotic stewardship programs to soon follow suit as well.
In 2022, a systematic analysis was published entitled “The Global Burden of Bacterial Antimicrobial Resistance in 2019.”12 This publication provided the first global estimates of the burden of bacterial AMR and the most comprehensive estimates to date. The systematic review included a broad range of pathogens and pathogen-drug combinations. The findings estimated that 4.95 million deaths were associated with bacterial AMR, which included 1.27 million deaths attributable to bacterial AMR.12 Estimations of deaths and disability-adjusted life-years were determined for 23 pathogens, 88 pathogen-drug combinations, and included 204 countries and territories.
Three infectious syndromes accounted for 79% of global AMR-associated deaths: lower respiratory tract infections (LRTI), bloodstream infections (BSI), and intra-abdominal infections (IAAI). Six pathogens accounted for 72% of Global AMR-associated deaths: E. coli, S. aureus, K. pneumoniae, S. pneumoniae, A. baumannii, and P. aeruginosa. Resistance to beta-lactams and fluoroquinolones accounted for greater than 70% of Global AMR-associated death.12 Seven pathogen-drug related combinations account for >50,000 deaths, including Methicillin-resistant S. aureus (MRSA) [>100,000 deaths], multi-drug resistant (MDR) M. tuberculosis, 3rd generation cephalosporin-resistant E. coli, carbapenem-resistant A. baumannii, fluroquinolone-resistant E. coli, carbapenem-resistant K. pneumoniae, and 3rd generation cephalosporin-resistant K. pneumoniae.
The top three global infectious disease threats in 2019 were AMR, (1.27M), malaria (696K), and HIV/AIDS (690K).12 Antimicrobial resistance was noted as the highest-burden in low-resource settings. Several goals were identified to combat rising AMR rates moving forward:
· Infection prevention and control to avoid the spread of AMR
· Vaccinations to reduce the need for antibiotics
· Improve access to essential antibiotics where needed
· Reduce exposure to antibiotics unrelated to human disease: One Health
· Optimize the use of antibiotics (antibiotic stewardship) guided by diagnostics (“building infrastructure that allows clinicians to diagnose infection accurately and rapidly is crucial”)
· Maintain investment in the development of a pipeline for new antibiotics
· Integrate fighting AMR as a priority in national strategies
· Increase microbiological laboratory and data collection capacity to improve data collection and understanding the threat of AMR
Several key points identified in the Global Burden of Bacterial Antimicrobial Resistance in 2019 follow a common theme expressed by Alain Mérieux in 2017: “Without diagnostics, medicine is blind.”13
The evolution of microbial testing has evolved over centuries and is currently advancing rapidly with automation, computerization, and nanotechnology. The first major discovery in microbiology was made in 1674 by Anthony van Leeuwenhoek when he peered through a drop of lake water through a glass magnifying lens he carefully ground.14 Although his lenses only magnified up to 300-fold, he was able to describe the three major shapes of bacteria: the cocci, bacilli, and spirilla. Robert Koch is credited for developing the pure culture techniques we use today. In the late 1870s, Koch realized that the isolation of pure cultures would be simplified on a solid medium on which a single isolated cell could multiply in a defined area. Koch’s laboratory also developed the Petri dish and using agar.14
Conventional blood culture processes and systems remained in place until the late 1960s when automated systems were introduced.15 Critical factors and recommended guidelines for optimal recovery of pathogens in blood were defined and established during the latter part of the 20th century and helped to develop best practices for the collection, processing, and interpretation of blood cultures. Examples of such critical factors and guidelines include but are not limited to, adequate skin disinfection, volume of blood collected, number and timing of blood cultures, blood culture bottles/media types, and duration of incubation and testing.16 With the establishment of critical factors came the need for better technology to process, monitor, and report on cultures and their results. Continuous blood culture monitoring systems were introduced and led to increased capacity of culture bottles, less manipulation, and earlier reporting of test results.15 The first automated blood culture systems were introduced in the early 1970s. With newer and faster technologies, the isolated rank order of recovery was also determined, which helped to provide input on automated testing development.15 DNA sequencing was developed in the late 1970s followed a decade later by the polymerase chain reaction (PCR) technology.21 These technologies are referred to as nucleic acid amplification tests (NAATs).17 PCR testing has evolved to include multiplex PCR (including and detecting multiple pathogens on a panel at once) and can provide results in about one hour.18
Applying automated diagnostic results to active antimicrobial stewardship has been shown to improve targeted antimicrobial therapy, improve patient care, and reduce antibiotic and length of stay costs (Figure 3).19 Integration of antimicrobial and diagnostic stewardship programs has also been an important step in the fight against AMR.20 Recently, a more robust multiplex PCR system was launched. It is FDA-approved and CLIA-waived with a smaller footprint and faster results turn-around time of about 15 minutes.21
The major advancements and improvements in molecular biology that were transitionally incorporated into sequencing technologies led to the second and third sequencing methodologies, commonly termed next-generation sequencing (NGS).17 The advantages of NGS compared with traditional sequencing methods include higher throughput (including multiplexing), higher sensitivity in detecting low-frequency variants, faster turnaround time for high sample volumes, and lower cost.22 Although NGS is not without limitations, it serves as a dramatic improvement in evaluating rare diseases, pathogen identification and antibiotic resistance profiles, and disease outbreak tracking in clinical settings (e.g., Ebola Virus, Malaria, and SARS-CoV-2).
Bacteriophage (phage) therapy has gained resurgent interest in recent years due to the lack of therapeutic options for patients unresponsive to conventional antimicrobials because of antimicrobial resistance.23 Phage therapy is considered compassionate, salvage therapy for patients with resistant pathogens from chronic/ recurrent infections such as urinary tract, respiratory, and skin/soft tissue infections. Other situations are implantable devices and joint replacements. Phage therapy is not FDA-approved and there is a need for more homogeneous randomized controlled trials to establish its place in patient care.23
Vaccines are a major achievement in medicine, but the development of more effective vaccines against infectious diseases is essential for the prevention and control of emerging pathogens worldide.24 Traditional vaccines, typically inactivated pathogens, have shown great success in the prevention or eradication of more than 30 infectious diseases.25
Additionally, mRNA vaccines have emerged as a revolution in vaccine fields due to their simplicity and adaptability in antigen design, the potential to induce both humoral and cell-mediated immune responses, and the high efficiency, and rapid, low-cost production in using similar manufacturing platforms for different mRNA vaccines. Recent research in infectious disease includes emerging or reemerging infectious pathogens (e.g., HIV, respiratory syncytial virus, influenza, streptococcus, etc.). Cancer vaccine research is in phase I/II clinical trials (e.g., colorectal, glioblastoma, ovarian, prostate, etc.).25
In conclusion, with rising rates of AMR and a lack of de novo antibiotics, maximizing our diagnostic capabilities is critical to use rapid and targeted therapy, in addition to phage and vaccine therapies to optimize patient care.
References
1. Will the virus have the last word? Office for Science and Society. Accessed May 22, 2024. https://www.mcgill.ca/oss/article/covid-19-history/will-virus-have-last-word.
2. The American association of immunologists - Alexander Fleming. Aai.org. Accessed May 22, 2024. https://www.aai.org/About/History/Notable-Members/Nobel-Laureates/AlexanderFleming.
3. Hutchings MI, Truman AW, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51:72-80. doi:10.1016/j.mib.2019.10.008.
4. Shi Z, Zhang J, Tian L, et al. A Comprehensive Overview of the Antibiotics Approved in the Last Two Decades: Retrospects and Prospects. Molecules. 2023;13;28(4):1762. doi:10.3390/molecules28041762.
5. President’s Council of advisors on science and technology. The White House. Published September 22, 2021. Accessed May 22, 2024. https://www.whitehouse.gov/pcast/.
6. Dellit TH, Owens RC, McGowan JE Jr, et al. Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis. 2007;15;44(2):159-77. doi:10.1086/510393.
7. Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2013. Atlanta: CDC, 2013.
8. Williams M, Kuper K. Overview of regulatory issues in antimicrobial stewardship. Infectious Diseases Pharmacotherapy Preparatory Review Course. Published online 2018.
9. Assistant Secretary for Health (ASH). Presidential advisory council on combating antibiotic-resistant bacteria (PACCARB). Hhs.gov. Published March 28, 2016. Accessed May 22, 2024. https://www.hhs.gov/ash/advisory-committees/paccarb/index.html.
10. Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an Antibiotic Stewardship Program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016;15;62(10):e51-77. doi:10.1093/cid/ciw118.
11. Cdc.gov. Accessed May 22, 2024. https://www.cdc.gov/antibiotic-use/data/outpatient-prescribing/index.html#.
12. Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;12;399(10325):629-655. doi:10.1016/S0140-6736(21)02724-0.
13. Mérieux A, President’s message, Institute Mérieux 2017.
14. Moore SA, Gery J. Fare m, Ed. The Microbial Perspective. CBS College Publishing; 1982.
15. Wilson ML. Development of new methods for detecting bloodstream pathogens. Clin Microbiol Infect. 2020;26(3):319-324. doi:10.1016/j.cmi.2019.08.002.
16. Wilson ML. Critical factors in the recovery of pathogenic microorganisms in blood. Clin Microbiol Infect. 2020;26(2):174-179. doi:10.1016/j.cmi.2019.07.023.
17. Vella S. The clinical value of next-generation sequencing integration within medical laboratories. Medical Laboratory Observer. Published May 18, 2022. Accessed May 22, 2024. https://www.mlo-online.com/disease/infectious-disease/article/21268056/the-clinical-value-of-next-generation-sequencing-integration-within-medical-laboratories.
18. Documents. BioFire Diagnostics. Published March 17, 2016. Accessed May 22, 2024. https://www.biofiredx.com/support/documents/.
19. Banerjee R, Teng CB, Cunningham SA, et al. Randomized Trial of Rapid Multiplex Polymerase Chain Reaction-Based Blood Culture Identification and Susceptibility Testing. Clin Infect Dis. 2015;1;61(7):1071-80. doi:10.1093/cid/civ447.
20. Patel R, Fang FC. Diagnostic Stewardship: Opportunity for a Laboratory-Infectious Diseases Partnership. Clin Infect Dis. 2018;16;67(5):799-801. doi:10.1093/cid/ciy077.
21. US FDA 510(k) Clearance and CLIA-waiver for the fast and innovative BIOFIRE® SPOTFIRE® System. bioMérieux Website. Accessed May 22, 2024. https://www.biomerieux.com/corp/en/journalists/press-releases/fda-clearance-clia-waiver-biofire-spotfire-bioMerieux.html.
22. Zhong Y, Xu F, Wu J, et al. Application of Next Generation Sequencing in Laboratory Medicine. Ann Lab Med. 2021;41(1):25-43. doi:10.3343/alm.2021.41.1.25.
23. Suh GA, Lodise TP, Tamma PD, et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob Agents Chemother. 2022;66(3):e0207121. doi:10.1128/AAC.02071-21.
24. Cable J, Graham BS, Koup RA, et al. Progress in vaccine development for infectious diseases-a Keystone Symposia report. Ann N Y Acad Sci. 2023;1524(1):65-86. doi:10.1111/nyas.14975.
25. Chen J, Chen J, Xu Q. Current Developments and Challenges of mRNA Vaccines. Annu Rev Biomed Eng. 2022;6;24:85-109. doi:10.1146/annurev-bioeng-110220-031722.
To take the test online go HERE. For more information, visit the Continuing Education tab.