Accurate detection of single nucleotide polymorphisms (SNPs) is important for many applications in the clinical laboratory, including diagnosis of disease and identification of appropriate therapeutic intervention based on specific genotypes. Quantitative real-time PCR (qPCR), in particular 5‘ nuclease assays such as qPCR assays, is commonly used for SNP genotyping as it allows accurate, high-throughput, and inexpensive genotyping from very little sample material.
However, while these assays are usually highly specific, not all targeted regions are optimal for PCR primer annealing or amplification. Common complications include primer-dimer formation—which consumes reagents and reduces target amplification and signal levels—and misamplification of very similar, off-target sequences. In SNP genotyping, these events can lead to false negative or false positive calls, ultimately complicating analyses and reducing confidence in results.
RNase H2–dependent PCR (rhPCR) is a novel method that significantly improves PCR specificity. Here we explain how rhPCR works, how it can be applied to alleviate problems in SNP genotyping, and the benefits it provides.
The beauty of blocking: rhPCR
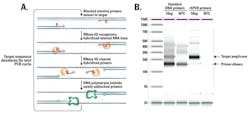
rhPCR was developed to increase specificity and accuracy of traditional PCR methods for a variety of challenging applications, including high-level multiplex assays and rare allele detection. The method uses a thermophilic RNase H2 enzyme from Pyrococcus abyssi, which functions optimally in conditions compatible with thermophilic DNA polymerases, such as Taq polymerase. This means RNase H2 can be integrated into PCR protocols easily and cost-effectively, with little to no modification of reaction temperatures, cycling times, or analysis procedures.
rhPCR primers provide internal checkpoints in the PCR reaction that improve specificity and prevent primer-dimers from depleting reagents. There are two key design elements that help them do this. First, rhPCR primers contain a terminal 3‘ modification that blocks extension. Second, these primers are unique in that they are comprised of DNA bases, except for a single RNA base close to their 3‘ ends. Once a primer binds to the target region, RNase H2 specifically recognizes and cleaves the RNA-DNA base pair, removing the RNA base and the terminal blocking group (Figure 1A).1
Primer unblocking by RNase H2 is very sensitive to correct base pairing and will be inhibited if there is a base mismatch in the cleavage site area. RNase H2 leaves an intact, 3‘ hydroxyl group available for extension by a DNA polymerase. However, extension occurs only when the primer has bound the matched target, ensuring any imperfect hybridization dimers will not amplify. This specificity of the RNase H2 enzyme minimizes subsequent off-target amplification of closely related sequences by DNA polymerase.2,3 Together, these characteristics prevent non-specific reactions, even in multiplex assays (Figure 1B).
Improving SNP genotyping with rhPCR
Traditional 5‘ nuclease PCR assays used for SNP genotyping rely on strong, detectable fluorescent signals. Therefore, any factors that reduce signal intensity will also reduce confidence and accuracy in genotyping calls. Both non-specific amplification and primer-dimer formation can reduce signal levels, leading to difficult-to-interpret or even unusable data. So, can the advantages of rhPCR minimize these issues and increase accuracy in SNP genotyping assays?
As discussed, in rhPCR, a mismatch at or near the RNA base inhibits the ability of RNase H2 to unblock an rhPCR primer. This unique property can be used as a basis for accurate SNP identification—the additional specificity requirements ensure only correct allelic matches are amplified to produce signal. Such assays have been developed by scientists and yield excellent results.
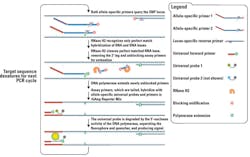
In these commercially-available, rhPCR-based SNP genotyping assays, two allele-specific rhPCR primers are used to query the SNP locus (Figure 2). The RNase H2 enzyme cleaves only the primer that is exactly matched to the target sequence, unblocking that specific primer for subsequent extension by polymerase. In this case a novel, mismatch-sensitive mutant Taq polymerase is used, further enhancing the specificity of the reaction.
This design provides additional value by incorporating a “tail“ on each allele-specific primer. The tail sequence is recognized by a universal probe-based reporter system. Use of a universal reporter system removes the need for the expensive, allele-specific, fluorescently labeled probes required for 5‘ nuclease assays. This significantly reduces costs. Signal is generated by polymerase extension causing release of the fluorophore from its nearby quencher. Importantly, the reporter probes only recognize the complement of the primer tail created during amplification. This means that the reporter probes cannot interact directly with assay primers, reducing non-specific background signal noise.
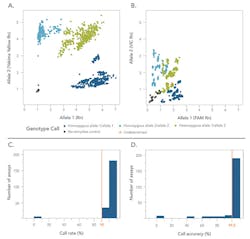
These combined features significantly boost signal-to-noise ratios compared with traditional 5‘ nuclease PCR assays (Figures 3A, 3B) and deliver calls with >99.5 percent accuracy (Figures 3C, 3D). The result is data that is much easier to interpret with confidence.
Reviewing the benefits
Accuracy, reliability, and confidence in results are essential in a clinical setting. Here, we have described how elegant modifications to 5‘ nuclease PCR chemistry offered by the rhPCR method can provide significant improvements to data quality and can be easily incorporated into existing workflows. These improvements facilitate a host of challenging PCR applications, including boosting specificity for rare allele detection in a background of wild-type DNA (such as in liquid biopsies), and in highly multiplexed amplification studies, where preventing primer-dimers from depleting reagents is particularly important.
The advantages of the rhPCR approach lend themselves particularly well to improving SNP genotyping. The combination of blocked-cleavable rhPCR primers, an enhanced Taq polymerase, and a universal reporter system significantly increase specificity and boost signal, enabling the method to very accurately discriminate among extremely similar sequences.
These rhPCR-based SNP genotyping assays effectively circumvent common issues in 5‘ nuclease assays, such as misamplification and primer-dimer formation. They deliver improved specificity and signal-to-noise ratio, increasing accuracy to >99.5 percent. This increased accuracy facilitates the ease of data interpretation and confidence in results that are so important in genotyping experiments.
REFERENCES
- Dobosy JR, Rose SD, Beltz K, et al. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable
primers. BMC Biotechnology. 2011;11:80. doi: 10.1186/1472-6750-11-80. - Prediger E. Discriminating highly similar transcripts using rhPCR. DECODED online. 2014. http://www.idtdna.com/pages/decoded/decoded-articles/core-concepts/decoded/2014/06/03/discriminating-highly-similar-transcripts-using-rhpcr.
- Boucard AA, Maxeiner S, Sudhof TC. Latrophilins function as heterophilic
cell-adhesion molecules by binding to teneurins: regulation by alternative splicing.
J Biol Chem. 2014 289(1):387–402. doi: 10.1074/jbc.M113.504779.
Aurita Menezes, PhD, serves as qPCR Product Manager for Integrated DNA Technologies, provider of rhPCR-based rhAmp SNP Assays and rhPCR reagents. She is responsible for training customer care representatives and technical and sales support representatives on the basics of PCR and qPCR, as well as supporting researchers in design, analysis, and interpretation of their qPCR data.